In 1952, Stanley Miller and Harold Urey carried out a chemical experiment to demonstrate how organic molecules could have formed spontaneously from inorganic precursors under prebiotic conditions like those posited by the Oparin–Haldane hypothesis. It used a highly reducing (lacking oxygen) mixture of gases—methane, ammonia, and hydrogen, as well as water vapor—to form simple organic monomers such as amino acids. Bernal said of the Miller–Urey experiment that "it is not enough to explain the formation of such molecules, what is necessary, is a physical-chemical explanation of the origins of these molecules that suggests the presence of suitable sources and sinks for free energy. However, current scientific consensus describes the primitive atmosphere as weakly reducing or neutral, diminishing the amount and variety of amino acids that could be produced. The addition of iron and carbonate minerals, present in early oceans, however produces a diverse array of amino acids. Later work has focused on two other potential reducing environments: outer space and deep-sea hydrothermal vents.
Stages in the origin of life range from the well-understood, such as the habitable Earth and the abiotic synthesis of simple molecules, to the largely unknown, like the derivation of the Last Universal Common Ancestor (LUCA) with its complex molecular functionalities.[1]
Life first- emerged at least 3.8 billion years ago (?), approximately 750 million years after Earth was formed (Figure above). How life originated and how the first cell came into being are matters of speculation, since these events cannot be reproduced in the laboratory.
Early universe with first stars:
Soon after the Big Bang, which occurred roughly 14 GA, (A= Billion years or Giga-annum (109 years), the only chemical elements present in the universe were Hydrogen, Helium, and Lithium, the three lightest atoms in the periodic table. These elements gradually came together to form stars. These early stars were massive and short-lived, producing all the heavier elements through stellar nucleosynthesis. Carbon, currently the fourth most abundant chemical elements in the universe (after hydrogen, helium and oxygen), was formed mainly in white dwarf stars, particularly those bigger than twice the mass of the sun. As these stars reached the end of their lifecycles, they ejected these heavier elements, among them carbon and oxygen, throughout the universe. These heavier elements allowed for the formation of new objects, including rocky planets and other bodies. According to the nebular hypothesis, the formation and evolution of the Solar System began 4.6 GA with the gravitational collapse of a small part of a giant molecular cloud. Most of the collapsing mass collected in the center, forming the Sun, while the rest flattened into a protoplanetary disk out of which the planets, moons, asteroids, and other small Solar System bodies formed.
The Earth was formed 4.54 GA. The Hadean Earth (from its formation until 4 GA) was at first inhospitable to any living organisms. During its formation, the Earth lost a significant part of its initial mass, and consequentially lacked the gravity to hold molecular hydrogen and the bulk of the original inert gases. The atmosphere consisted largely of water vapor, nitrogen, and carbon dioxide, with smaller amounts of carbon monoxide, hydrogen, and sulfur compounds.[55] The solution of carbon dioxide in water is thought to have made the seas slightly acidic, with a pH of about 5.5. The Hadean atmosphere has been characterized as a "gigantic, productive outdoor chemical laboratory, similar to volcanic gases today which still support some abiotic chemistry. Earliest evidence of life.
Main article: Earliest known life forms
Life existed on Earth more than 3.5 GA, during the Eoarchean when sufficient crust had solidified following the molten Hadean. The earliest physical evidence of life so far found consists of microfossils in the Nuvvuagittuq Greenstone Belt of Northern Quebec, in banded iron formation rocks at least 3.77 and possibly 4.28 GA. The micro-organisms lived within hydrothermal vent precipitates, soon after the 4.4 GA formation of oceans during the Hadean. The microbes resembled modern hydrothermal vent bacteria, supporting the view that abiogenesis began in such an environment.
Biogenic graphite has been found in 3.7 GA metasedimentary rocks from southwestern Greenland and in microbial mat fossils from 3.49 GA Western Australian sandstone. Evidence of early life in rocks from Akilia Island, near the Isua supracrustal belt in southwestern Greenland, dating to 3.7 GA, have shown biogenic carbon isotopes.[78] In other parts of the Isua supracrustal belt, graphite inclusions trapped within garnet crystals are connected to the other elements of life: oxygen, nitrogen, and possibly phosphorus in the form of phosphate, providing further evidence for life 3.7 GA. In the Pilbara region of Western Australia, compelling evidence of early life was found in pyrite-bearing sandstone in a fossilized beach, with rounded tubular cells that oxidized sulfur by photosynthesis in the absence of oxygen. Zircons from Western Australia imply that life existed on Earth at least 4.1 GA years back , GA means 109 yrs. The Pilbara region of Western Australia contains the Dresser Formation with rocks 3.48 GA, including layered structures called stromatolites. Their modern counterparts are created by photosynthetic micro-organisms including cyanobacteria.[83] These lie within undeformed hydrothermal-sedimentary strata; their texture indicates a biogenic origin. Parts of the Dresser formation preserve hot springs on land, but other regions seem to have been shallow seas.
Stromatolites in the Siyeh Formation, Glacier National Park, dated 3.5 GA, placing them among the earliest life-forms
Modern stromatolites in Shark Bay, created by photosynthetic cyanobacteria.
All chemical elements except for hydrogen and helium derive from stellar nucleosynthesis. The basic chemical ingredients of life – the carbon-hydrogen molecule (CH), the carbon-hydrogen positive ion (CH+) and the carbon ion (C+) – were produced by ultraviolet light from stars.[85] Complex molecules, including organic molecules, form naturally both in space and on planets.[86] Organic molecules on the early Earth could have had either terrestrial origins, with organic molecule synthesis driven by impact shocks or by other energy sources, such as ultraviolet light, redox coupling, or electrical discharges; or extraterrestrial origins (pseudo-panspermia), with organic molecules formed in interstellar dust clouds raining down on to the planet.
Nucleobases.
The majority of organic compounds introduced on Earth by interstellar dust particles have helped to form complex molecules, thanks to their peculiar surface-catalytic activities. Studies of the 12C/13C -isotopic ratios of organic compounds in the Murchison meteorite suggest that the RNA component uracil and related molecules, including xanthine, were formed extra terrestrially. NASA studies of meteorites suggest that all four DNA nucleobases (adenine, guanine and related organic molecules) have been formed in outer space. The cosmic dust permeating the universe contains complex organics ("amorphous organic solids with a mixed aromatic–aliphatic structure") that could be created rapidly by stars. Glycolaldehyde, a sugar molecule and RNA precursor, has been detected in regions of space including around protostars and on meteorites.
The lipid world theory postulates that the first self-replicating object was lipid-like. Phospholipids form lipid bilayers in water while under agitation—the same structure as in cell membranes. These molecules were not present on early Earth, but other amphiphilic long-chain molecules also form membranes. These bodies may expand by insertion of additional lipids, and may spontaneously split into two offspring of similar size and composition. The main idea is that the molecular composition of the lipid bodies is a preliminary to information storage, and that evolution led to the appearance of polymers like RNA that store information. Studies on vesicles from amphiphiles that might have existed in the prebiotic world have so far been limited to systems of one or two types of amphiphiles.
A lipid bilayer membrane could be composed of a huge number of combinations of arrangements of amphiphiles. The best of these would have favored the constitution of a hypercycle, actually a positive feedback composed of two mutual catalysts represented by a membrane site and a specific compound trapped in the vesicle. Such site/compound pairs are transmissible to the daughter vesicles leading to the emergence of distinct lineages of vesicles, which would have allowed natural selection.
Chemiosmosis.
ATP synthase uses the chemiosmotic proton gradient to power ATP synthesis through oxidative phosphorylation.
In 1961, Peter Mitchell proposed chemiosmosis as a cell's primary system of energy conversion. The mechanism, now ubiquitous in living cells, powers energy conversion in micro-organisms and in the mitochondria of eukaryotes, making it a likely candidate for early life.[149][150] Mitochondria produce adenosine triphosphate (ATP), the energy currency of the cell used to drive cellular processes such as chemical syntheses. The mechanism of ATP synthesis involves a closed membrane in which the ATP synthase enzyme is embedded. The energy required to release strongly-bound ATP has its origin in protons that move across the membrane. In modern cells, those proton movements are caused by the pumping of ions across the membrane, maintaining an electrochemical gradient. In the first organisms, the gradient could have been provided by the difference in chemical composition between the flow from a hydrothermal vent and the surrounding seawater or perhaps meteoric quinones that were conducive to the development of chemiosmotic energy across lipid membranes if at a terrestrial origin.
Chemiosmotic coupling in the membranes of a mitochondrion
The RNA world.
Main article: RNA world.
The RNA world hypothesis describes an early Earth formation of self-replicating and catalytic RNA and not DNA or proteins. Many researchers concur that an RNA world must have preceded the DNA-based life that now dominates. However, RNA-based life may not have been the first to exist. Another model echoes Darwin's "warm little pond" with cycles of wetting and drying.
RNA is central to the translation process. Small RNAs can catalyze all the chemical groups and information transfers required for life. RNA both expresses and maintains genetic information in modern organisms; and the chemical components of RNA are easily synthesized under the conditions that approximated the early Earth, which were very different from those that prevail today. The structure of the ribozyme has been called the "smoking gun", with a central core of RNA and no amino acid side chains within 18 Ĺ of the active site that catalyzes peptide bond formation.
The concept of the RNA world was proposed in 1962 by Alexander Rich, and the term was coined by Walter Gilbert in 1986. There were initial difficulties in the explanation of the abiotic synthesis of the nucleotides cytosine and uracil. Subsequent research has shown possible routes of synthesis; for example, formamide produces all four ribonucleotides and other biological molecules when warmed in the presence of various terrestrial minerals.
The RNA world hypothesis proposes that undirected polymerization led to the emergence of ribozymes, and in turn to an RNA replicase.
RNA replicase can function as both coded and catalyst for further RNA replication, i.e. it can be autocatalytic. Jack Szostak has shown that certain catalytic RNAs can join smaller RNA sequences together, creating the potential for self-replication. The RNA replication systems, which include two ribozymes that catalyze each other's synthesis, showed a doubling time of the product of about one hour, and were subject to natural selection under the experimental conditions. If such conditions were present on early Earth, then natural selection would favor the proliferation of such autocatalytic sets, to which further functionalities could be added. Self-assembly of RNA may occur spontaneously in hydrothermal vents. A preliminary form of tRNA could have assembled into such a replicator molecule.
Possible precursors to protein synthesis include the synthesis of short peptide cofactors or the self-catalyzing duplication of RNA. It is likely that the ancestral ribosome was composed entirely of RNA, although some roles have since been taken over by proteins. Major remaining questions on this topic include identifying the selective force for the evolution of the ribosome and determining how the genetic code arose.
Eugene Koonin has argued that "no compelling scenarios currently exist for the origin of replication and translation, the key processes that together comprise the core of biological systems and the apparent pre-requisite of biological evolution. The RNA World concept might offer the best chance for the resolution of this conundrum but so far cannot adequately account for the emergence of an efficient RNA replicase or the translation system."
Phylogeny and LUCA[edit]
Further information: Last universal common ancestor.
Starting with the work of Carl Woes from 1977 onwards, genomics studies have placed the Last Universal Common Ancestor (LUCA) of all modern life-forms between Bacteria and a clade formed by Archaea and Eukaryota in the phylogenetic tree of life. It lived over 4 Gya. A minority of studies have placed the LUCA in Bacteria, proposing that Archaea and Eukaryota are evolutionarily derived from within Eubacteria; Thomas Cavalier-Smith suggested that the phenotypically diverse bacterial phylum Chloroflexota contained the LUCA.
Phylogeny and LUCA.
Further information: Last universal common ancestor
Starting with the work of Carl Woes from 1977 onwards, genomics studies have placed the last universal common ancestor (LUCA) of all modern life-forms between Bacteria and a clade formed by Archaea and Eukaryote in the phylogenetic tree of life. It lived over 4 GA. A minority of studies have placed the LUCA in Bacteria, proposing that Archaea and Eukaryota are evolutionarily derived from within Eubacteria; Thomas Cavalier-Smith suggested that the phenotypically diverse bacterial phylum Chloroflexota contained the LUCA.
Phylogenetic tree above showing the Last Universal Common Ancestor (LUCA) at the root. The major clades are the Bacteria on one hand, and the Archaea and Eukaryota on the other.
In 2016, a set of 355 genes likely present in the LUCA was identified. A total of 6.1 million prokaryotic genes from Bacteria and Archaea were sequenced, identifying 355 protein clusters from amongst 286,514 protein clusters that were probably common to the LUCA. The results suggest that the LUCA was anaerobic with a Wood–Ljungdahl pathway, nitrogen- and carbon-fixing, thermophilic. Its cofactors suggest dependence upon an environment rich in hydrogen, carbon dioxide, iron, and transition metals. Its genetic material was probably DNA, requiring the 4-nucleotide genetic code, messenger RNA, transfer RNAs, and ribosomes to translate the code into proteins such as enzymes. LUCA likely inhabited an anaerobic hydrothermal vent setting in a geochemically active environment. It was evidently already a complex organism, and must have had precursors; it was not the first living thing. The physiology of LUCA has been in dispute.
Leslie Orgel argued that early translation machinery for the genetic code would be susceptible to error catastrophe. Geoffrey Hoffmann however showed that such machinery can be stable in function against "Orgel's paradox".
Water- single celled life:
We may never be able to prove beyond any doubt how life first evolved. But of the many explanations proposed, one stands out – the idea that life evolved in hydrothermal vents deep under the sea. Not in the superhot black smokers, but more placid affairs known as alkaline hydrothermal vents.
This theory can explain life’s strangest feature, and there is growing evidence to support it.
Earlier this year, for instance, lab experiments confirmed that conditions in some of the numerous pores within the vents can lead to high concentrations of large molecules. This makes the vents an ideal setting for the “RNA world” widely thought to have preceded the first cells.
If life did evolve in alkaline hydrothermal vents, it might have happened something like this:
1.
Water percolated down into newly formed rock under the seafloor, where it reacted with minerals such as olivine, producing a warm alkaline fluid rich in hydrogen, sulphides and other chemicals – a process called ‘serpentinization’.
This hot fluid welled up at alkaline hydrothermal vents like those at the Lost City, a vent system discovered near the Mid-Atlantic Ridge in 2000.
2.
Unlike today’s seas, the early ocean was acidic and rich in dissolved iron. When upwelling hydrothermal fluids reacted with this primordial seawater, they produced carbonate rocks riddled with tiny pores and a “foam” of iron-Sulphur bubbles.
3.
Inside the iron-Sulphur bubbles, hydrogen reacted with carbon dioxide, forming simple organic molecules such as methane, formate and acetate. Some of these reactions were catalyzed by the iron-Sulphur minerals. Similar iron-Sulphur catalysts are still found at the heart of many proteins today.
4.
The electrochemical gradient between the alkaline vent fluid and the acidic seawater leads to the spontaneous formation of acetyl phosphate and pyrophosphate, which act just like adenosine triphosphate or ATP, the chemical that powers living cells.
These molecules drove to the formation of amino acids – the building blocks of proteins – and nucleotides-the building blocks for RNA and DNA.
5.
Thermal currents and diffusion within the vent pores concentrated larger molecules like nucleotides, driving the formation of RNA and DNA – and providing an ideal setting for their evolution into the world of DNA and proteins. Evolution got under way, with sets of molecules capable of producing more of themselves starting to dominate.
6.
Fatty molecules coated the iron-Sulphur froth and spontaneously formed cell-like bubbles. Some of these bubbles would have enclosed self-replicating sets of molecules – the first organic cells. The earliest protocells may have been elusive entities, though, often dissolving and reforming as they circulated within the vents.
7.
The evolution of an enzyme called pyrophosphatase, which catalyzes the production of pyrophosphate, allowed the protocells to extract more energy from the gradient between the alkaline vent fluid and the acidic ocean. This ancient enzyme is still found in many bacteria and archaea, the first two branches on the tree of life.
8.
Some protocells started using ATP as well as acetyl phosphate and pyrophosphate. The production of ATP using energy from the electrochemical gradient is perfected with the evolution of the enzyme ATP synthase, found within all life today.
9.
Protocells further from the main vent axis, where the natural electrochemical gradient is weaker, started to generate their own gradient by pumping protons across their membranes, using the energy released when carbon dioxide reacts with hydrogen.
This reaction yields only a small amount of energy, not enough to make ATP. By repeating the reaction and storing the energy in the form of an electrochemical gradient, however, protocells “saved up” enough energy for ATP production.
10.
Once protocells could generate their own electrochemical gradient, they were no longer tied to the vents. Cells left the vents on two separate occasions, with one exodus giving rise to bacteria and the other to archaea.
Back to feature: Was our oldest ancestor a proton-powered rock? New scientist. https://www.newscientist.com/
What was needed for the first cell? Some sort of membrane surrounding organic molecules? Probably.
How, organic molecules such as RNA developed into cells is not known for certain. Scientists speculate that lipid membranes grew around the organic molecules. The membranes prevented the molecules from reacting with other molecules, so they did not form new compounds. In this way, the organic molecules persisted, and the first cells may have formed. Figure below shows a model of the hypothetical first cell. Were these first cells the first living organisms? Were they able to live and reproduce while passing their genetic information to the next generation? If so, then yes, these first cells could be considered the first living organisms.

Hypothetical First Cell. The earliest cells may have consisted of little more than RNA inside a lipid membrane.
LUCA.
No doubt there were many early cells of this type. However, scientists think that only one early cell (or group of cells) eventually gave rise to all subsequent life on Earth. That one cell is called the Last Universal Common Ancestor (LUCA). It probably existed around 3.5 billion years ago. LUCA was one of the earliest prokaryotic cells. It would have lacked a nucleus and other membrane-bound organelles. To learn more about LUCA and universal common descent, you can watch the video at the following link: http:// www.youtube.com/watch? v=G0UGpcea8Zg.
· The first cells consisted of little more than an organic molecule such as RNA inside a lipid membrane.
· One cell (or group of cells), called the last universal common ancestor (LUCA), gave rise to all subsequent life on Earth.
· Photosynthesis evolved by 3 billion years ago and released oxygen into the atmosphere.
· Cellular respiration evolved after that to make use of the oxygen.
LUCA
No doubt there were many early cells of this type. However, scientists think that only one early cell (or group of cells) eventually gave rise to all subsequent life on Earth. That one cell is called the Last Universal Common Ancestor (LUCA). It probably existed around 3.5 billion years ago. LUCA was one of the earliest prokaryotic cells. It would have lacked a nucleus and other membrane-bound organelles. To learn more about LUCA and universal common descent, you can watch the video at the following link: http://www.youtube.com/watch?v=G0UGpcea8Zg.
Photosynthesis and Cellular Respiration;
The earliest cells were probably heterotrophs. Most likely they got their energy from other molecules in that time “organic-soup.” However, by about 3 billion years ago, a new way of obtaining energy evolved. This new way was photosynthesis. Through photosynthesis, organisms could use sunlight to make food from carbon dioxide and water. These organisms were the first autotrophs. They provided food for themselves and for other organisms that began to consume them.
After photosynthesis evolved, oxygen started to accumulate in the atmosphere. This has been dubbed the “oxygen catastrophe.” Why? Oxygen was toxic to most early cells because they had evolved in its absence. As a result, many of them died out. The few that survived evolved a new way to take advantage of the oxygen. This second major innovation was cellular respiration. It allowed cells to use oxygen to obtain more energy from organic molecules.
Summary
· The first cells consisted of little more than an organic molecule such as RNA inside a lipid membrane.
· One cell (or group of cells), called the last universal common ancestor (LUCA), gave rise to all subsequent life on Earth.
· Photosynthesis evolved by 3 billion years ago and released oxygen into the atmosphere.
· Cellular respiration evolved after that to make use of the oxygen.
Formation life molecules- many-many million years since the origin of earth- production life required molecules and association of them using membrane forms; more than anything life as a molecular form coordinating from each other and heritable-it has molecular origin. that is the fundamental; to function the life molecule requires DNA and RNA and importantly polypeptide molecules and lipid molecules to enclose them all in one area.
The history of Earth concerns the development of planet Earth from its formation to the present day. Nearly all branches of natural science have contributed to understanding of the main events of Earth's past, characterized by constant geological change and biological evolution.

Early life may have emerged from a mixture of RNA and DNA building blocks, developing the two nucleic acids simultaneously instead of evolving DNA from RNA.https://www.scientificamerican.com/
As RNA gave way to DNA, some think a mixture of nucleotide building blocks would have been inevitable. As these nucleotides connected to form strands, the thermodynamic and kinetic stability of pure RNA and DNA duplexes would drive these nucleic acids to accumulate in primitive cells, while less thermally stable complexes containing one strand of RNA and one strand of DNA fell apart.

Research in different fields is coming together to assemble a more complete picture of the way the RNA World began and operated. https://www.nature.com/

Minor and Major Spliceosome Assembly and Catalytic Core Formation.
https://www.cell.com/

Figure; An RNA molecule that can catalyze its own synthesis
Figure; An RNA molecule that can catalyze its own synthesis
This hypothetical process would require catalysis of the production of both a second RNA strand of complementary nucleotide sequence and the use of this second RNA molecule as a template to form many molecules of RNA with the original sequence. The red rays represent the active site of this hypothetical RNA enzyme.
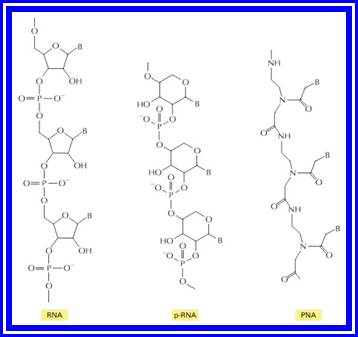
Figure: Structures of RNA from the left and two related information-carrying polymers; pRNA, and PNA;
In each case, B indicates the positions of purine and pyrimidine bases. The polymer p-RNA (pyranosyl-RNA) is RNA in which the furanose (five-membered ring) form of ribose has been replaced by the pyranose (six-membered ring) form. In PNA (peptide nucleic acid), the ribose phosphate backbone of RNA has been replaced by the peptide backbone found in proteins. Like RNA, both p-RNA and PNA can form double helices through complementary base-pairing, and each could therefore in principle serve as a template for its own synthesis.
Early life may have emerged from a mixture of RNA and DNA building blocks, developing the two nucleic acids simultaneously instead of evolving DNA from RNA.

Employing the module strategy based on our recent finding that the recognition behavior of peptide ribonucleic acid (PRNA) with complementary DNA/RNA is effectively controlled by the anti-to-syn orientation switching of pyrimidine nucleobase induced by borate ester formation, we designed and synthesized PRNA–DNA and PRNA–PNA–DNA chimeras. In these chimeras, both of the PRNA (or PRNA–PNA) and DNA domains recognize the complementary DNA/RNA to form a stable complex, and the PRNA domain is simultaneously expected to play the dual role of switching the recognition behavior and inhibiting hydrolysis by exonucleases. The complexation and recognition control behaviors of these chimeras with DNA and RNA have been elucidated. https://www.journal.csj.jp/

Beginning with a large pool of nucleic acid molecules synthesized in the laboratory, those rare RNA molecules that possess a specified catalytic activity can be isolated and studied. Although a specific example (that of an auto-phosphorylating -ribozyme) is shown, variations of this procedure have been used to generate many of the ribozymes listed. During the autophosphorylation step, the RNA molecules are sufficiently dilute to prevent the “cross”-phosphorylation of additional RNA molecules. In reality, several repetitions of this procedure are necessary to select the very rare RNA molecules with catalytic activity. Thus the material initially eluted from the column is converted back into DNA, amplified many fold (using reverse transcriptase and PCR as explained in Chapter 8), transcribed back into RNA, and subjected to repeated rounds of selection. (Adapted from J.R. Lorsch and J.W. Szostak, Nature 371:31–36, 1994.)
From our knowledge of present-day organisms and the molecules they contain, it seems likely that the development of the directly autocatalytic mechanisms fundamental to living systems began with the evolution of families of molecules that could catalyze their own replication. With time, a family of cooperating RNA catalysts probably developed the ability to direct synthesis of polypeptides. DNA is likely to have been a late addition: as the accumulation of additional protein catalysts allowed more efficient and complex cells to evolve, the DNA double helix replaced RNA as a more stable molecule for storing the increased amounts of genetic information required by such cells.
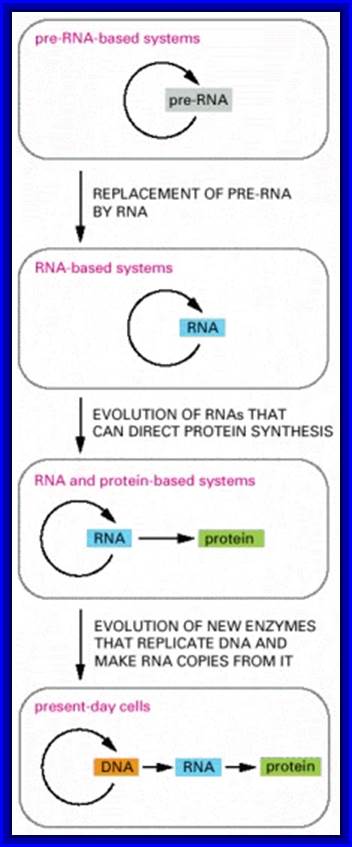
Figure- above. The hypothesis that RNA preceded DNA and proteins in evolution;
In the earliest cells, pre-RNA molecules would have had combined genetic, structural, and catalytic functions and these functions would have gradually been replaced by RNA. In present-day cells, DNA is the repository of genetic information, and proteins perform the vast majority of catalytic functions in cells. RNA primarily functions today as a go-between in protein synthesis, although it remains a catalyst for a number of crucial reactions.
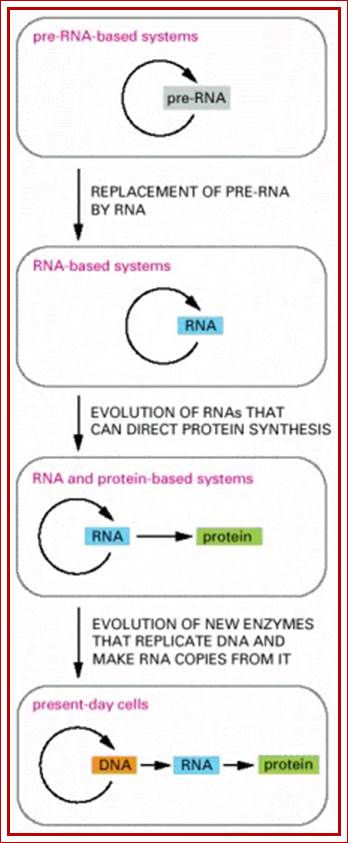
Fig; RNA to RNA; RNA to Proteins, DNA to RNA to Proteins; where is RNA to DNA to RNA?
Figure: The hypothesis that RNA preceded DNA and proteins in evolution.
In the earliest cells, Pre-RNA molecules would have had combined genetic, structural, and catalytic functions and these functions would have gradually been replaced by DNA. In present-day cells, DNA is the repository of genetic information, and proteins perform the vast majority of catalytic functions in cells. RNA primarily functions today as a go-between in protein synthesis, although it remains a catalyst for a number of crucial reactions.
Scientists find that an RNA-DNA mix may have created the first life on our planet. DNA model being created. New study shows that RNA and DNA likely originated together? The mixture of the acids is believed to have produced Earth's first life forms.
Experts now think that Miller and Urey’s experiments didn’t get the atmosphere of early Earth quite right, but more recent experiments have shown that organic building blocks, including nucleotides, can form under a relatively wide range of conditions that could have been present on the primordial earth.

Pathways for the synthesis of RNA and DNA
Figure (above).
Metabolic pathways for RNA and DNA precursors biosynthesis: a palimpsest from the RNA to DNA world transition? The biosynthetic pathways for purine and pyrimidine nucleotides both start with ribose 5-monophosphate. The formation of the four bases requires several amino-acids, formate and carbamoyl-phosphate. Nucleotide monophosphates (NMP) are converted into RNA precursors (NTP) by NMP kinases (k) and NDP kinases (K). These reactions probably are relics of the RNA-protein world. DNA precursors are produced from NDP and/or NTP by ribonucleotide reductases (RNR), except for dTTP, which results from methylation of dUMP. dTMP is produced from dUMP by Thymidylate synthases (ThyA or ThyX) and converted into dTTP by the same kinases that convert NMP into NTP. dUMP can be produced either by dUTPAse or by dCTP deaminase. In the U-DNA world, it could have been also produced by degradation of U-DNA. The mode of dTMP production clearly suggests that U-DNA was an evolutionary intermediate between RNA and T-DNA. Some viruses contain U-DNA, whereas others contain HMC-DNA (HMC= hydroxymethyl-cytosine). Transformation of C into HMC occurs at the level of dCMP, and conversion of dCMP into dHMCMP is catalyzed by a dCMP hydroxy-methyl transferase (dCMP HM transferase), which is homologue to ThyA (See refs. 11, 14, and 19 for more details).
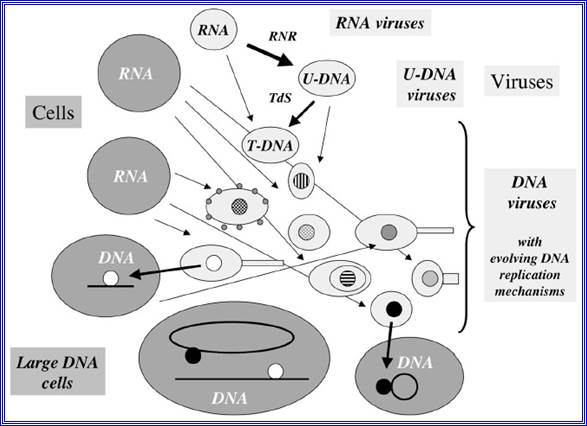
RNA to DNA
Figure- above:
Evolution of DNA replication mechanisms in the viral world? This figure illustrates a coevolution scenario of cells and viruses in the transition from the RNA to the DNA world. Large gray circles or ovals indicate cells, whereas small light grey circles ovals (some with tails) indicate viruses. In this scenario, different replication mechanisms (inner circles with different colors) originated among various viral lineages after the invention of U-DNA and T-DNA by viruses (RNR= ribonucleotide reductase, TdS=Thymidylate synthase).7 These mechanisms evolved through the independent recruitment of cellular or viral enzymes involved in RNA replication or transcription (polymerases, helicases, nucleotide binding proteins) to produce enzymes involved in DNA replication (thin arrows). Two different DNA replication mechanisms (black and white circles) were finally transferred independently to cells (thick arrows). These two transfers can have occurred either before or after the LUCA. In the first case, the two systems might have been present in LUCA via cell fusion or successive transfers. One system could have also replaced the other in a particular cell lineage (for these different possibilities, see fig.)

![]()
An artist's depiction of gas and dust in the forms planets, surrounding a young star in their orbits. (Image credit: NASA).
The solar system is anchored by our Sun. Before the solar system existed, a massive concentration of interstellar gas and dust created a molecular cloud that actually formed the sun's birthplace. Cold temperatures caused the gas to clump together, growing steadily denser. The densest parts of the cloud began to collapse under their own gravity, perhaps with a nudge from a nearby stellar explosion, forming a wealth of young stellar objects known as protostars.
Gravity continued to collapse the material onto the infant solar system, creating a star and a disk of material from which the planets would form. Eventually, the newborn sun encompassed more than 99% of the solar system's mass, according to NASA (opens in new tab). When pressure inside the star grew so powerful that fusion kicked in, turning hydrogen to helium, the star began to blast a stellar wind that helped clear out the debris and stopped it from falling inward.
Although gas and dust shroud young stars in visible wavelengths, infrared telescopes have probed many clouds in the Milky Way galaxy to study the environment of other newborn stars. Scientists have applied what they've seen in other systems to our own star.
![]()
An artist's depiction of gas and dust surrounding a young star. (Image credit: NASA). The solar system is anchored by our sun. Before the solar system existed, a massive concentration of interstellar gas and dust created a molecular cloud that would form the sun's birthplace. Cold temperatures caused the gas to clump together, growing steadily denser. The densest parts of the cloud began to collapse under their own rotational gravity, perhaps with a nudge from a nearby stellar explosion, forming a wealth of young stellar objects known as protostars.
Gravity continued to collapse the material onto the infant solar system, creating a star and a disk of material from which the planets would form. Eventually, the newborn sun encompassed more than 99% of the solar system's mass, according to NASA(opens in new tab). When pressure inside the star grew so powerful that fusion kicked in, turning hydrogen to helium, the star began to blast a stellar wind that helped clear out the debris and stopped it from falling inward.
Although gas and dust shroud young stars in visible wavelengths, infrared telescopes have probed many clouds in the Milky Way galaxy to study the environment of other newborn stars. Scientists have applied what they've seen in other systems to our own star.
![]()
An artist's depiction of gas and dust surrounding a young star. (Image credit: NASA). The solar system is anchored by our sun.
Before the solar system existed, a massive concentration of interstellar gas and dust created a molecular cloud that would form the sun's birthplace. Cold temperatures caused the gas to clump together, growing steadily denser. The densest parts of the cloud began to collapse under their own gravity, perhaps with a nudge from a nearby stellar explosion, forming a wealth of young stellar objects known as protostars.
Gravity continued to collapse the material onto the infant solar system, creating a star and a disk of material from which the planets would form. Eventually, the newborn sun encompassed more than 99% of the solar system's mass, according to NASA, (opens in new tab). When pressure inside the star grew so powerful that fusion kicked in, turning hydrogen to helium, the star began to blast a stellar wind that helped clear out the debris and stopped it from falling inward.
Although gas and dust shroud young stars in visible wavelengths, infrared telescopes have probed many clouds in the Milky Way galaxy to study the environment of other newborn stars. Scientists have applied what they've seen in other systems to our own star.

Living - Cell’s planets
water-attracting heads of lipid molecules
water-repellent heads
|
||||||||||||||||||||||||||||||||||
One possible family tree of eukaryotes -
Main article: Eukaryote
Chromatin, nucleus, endomembrane system, and mitochondria:]
Eukaryotes may have been present long before the oxygenation of the atmosphere, but most modern eukaryotes require oxygen, which is used by their mitochondria to fuel the production of ATP, the internal energy supply of all known cells. In the 1970s, a vigorous debate concluded that eukaryotes emerged as a result of a sequence of endosymbiosis between prokaryotes. For example: a predatory microorganism invaded a large prokaryote, probably an Archaean, but instead of killing its prey, the attacker took up residence and evolved into mitochondria; one of these chimeras later tried to swallow a photosynthesizing cyanobacterium, but the victim survived inside the attacker and the new combination became the ancestor of Plants; and so on. After each endosymbiosis, the partners eventually eliminated unproductive duplication of genetic functions by re-arranging their genomes, a process which sometimes involved transfer of genes between them. Another hypothesis proposes that mitochondria were originally sulfur- or hydrogen-metabolizing endosymbionts, and became oxygen-consumers later. On the other hand, mitochondria might have been part of eukaryotes' original equipment.
There is a debate about when eukaryotes first appeared: the presence of steranes in Australian shales may indicate eukaryotes at 2.7 GA; however, an analysis in 2008 concluded that these chemicals infiltrated the rocks less than 2.2 GA and prove nothing about the origins of eukaryotes. Fossils of the algae-Grypania have been reported in 1.85 billion-year-old rocks (originally dated to 2.1 GA but later revised), indicating that eukaryotes with organelles had already evolved. A diverse collection of fossil algae was found in rocks dated between 1.5 and 1.4 GA. The earliest known fossils of fungi date from 1.43 GA.
CELL_STRUCTURE:
Cell is the unit of structure and function. They are the building blocks of an organism. Irrespective of the nature of organisms (plant or animal) they are either made up of single cell or many cells, the former is called unicellular and later is called multicellular organisms; in the later cells are differentiated into various kinds and they are grouped into tissues, which perform special and special functions.
An Apple with such Sweet Cells.
![]()
![]()
![]()
![]()
Example meristematic cells perform repeated cell divisions, phloem cells- conduct food material, sclerenchyma- mechanical support function, xylem- conduction of water and mineral salts and so on. Nevertheless, all these different types of cells are derived from the same embryonic cells. The development of various cell types from a single cell is determined and regulated by a process called differentiation, which in turn is controlled by differential concentrations of plant hormones. Added to this, organ differentiation is another fascinating aspect of development. All these processes are regulated by differential gene regulation in response to environmental stimulus and phytohormones.
Cellular composition:
All cells are made up of a semi viscous fluid called protoplasm, which is considered as the physical basis of life, for it controls all biochemical reactions of the cell. In fact, it is the microcosm of life with many secrets not known to us. The protoplasm is colloidal in nature, because many cell colloidal sized structures and macromolecules are suspended in it. It also exhibits sol and gel properties. The granular nature of the protoplasm is due to the presence of many tiny organelles. Vacuoles of various types are also found, but in plant cells when it is matured, a large central vacuole is present and it is separated from the rest of the protoplasm by a single unit membrane called tonoplast. The fluid present is called cell sap. There are many common features between plant and animal cells; the former is distinguished by the presence of distinct cell wall and plastids, which are totally absent in animal cells. However, centrosomes are invariably present in animal cells and rarely in plant cells with exception of some lower plant unicellular algae like Chlamydomonas.
Chemical composition:
When cells are subjected to chemical analysis the following compounds are found (approximate value).
Compound Animal plant cell
Carbohydrates 20% 30%
Proteins 45% 40%
Lipids 30% 25%
RNA 1% 0.4%
DNA 0.2% 0.4%
Inorganic & others 3.8% 3.6%
Carbohydrates: These are organic compounds consisting of C, H and O. The basic structural components of carbohydrates are monosaccharide sugars consisting of 3 to 7 carbons. Example: Glucose (6c), Fructose (6c), Erythrose (4c), Xylose (5c), etc. such monomers by undergoing polymerization develop into long chained polysaccharides, such as cellulose, cellobioses, starch and glycogen (animal starch). Cellulose is made up of glucose units linked by β1>4 linkages and it is an important component of cell wall. Similarly, starch is also a polymer of α1-4 linked glucose units and it is the main source of energy for all living cells.
Proteins: proteins are the most important organic components of cells, for they act as structural as well as functional molecules. With out proteins life cannot exist, though DNA is the genetic material it is proteins that make it or break it; DNA is master library with information, that cannot be changed. Proteins are made up of basic building blocks called amino acids. The polymers of amino acid residues are called polypeptides (proteins) which exhibit different structural conformation (shape). The 3-D shapes are specific and characteristic for a particular protein, thus they exhibit specific structure and function. Exemple: contractile proteins (muscles), transport proteins (Hemoglobin), enzyme proteins, hormonal proteins (insulin), antibody proteins IgG etc. Almost all biochemical activities, including growth and development are controlled by proteins, without which cell ceases to live.
Lipids: Fatty acids and their derivatives are very important for two reasons, firstly lipids like lecithin, phosphotidyl ethanolamine, sphingolipids, glycolipids, steroids and others are part of the cellular membranes; thus, they contribute to the structure of the membranes. Secondly lipids also act as food reserve and provide energy by oxidation.
Nucleic acids: These are the polymers of nucleotides, consisting of nitrogenous bases, phosphates and pentose sugars. There are two types of nucleic acid – 1) Deoxy-ribose nucleic acid (DNA), 2) Ribose nucleic acid (RNA). DNA is mostly found in chromosomes; it is the repository of genetic information and it provides information for the synthesis of proteins. The chemical composition, structure and function will be discussed in the chapter elsewhere.
Inorganic/Organic factors: Many inorganic metals Fe, Mg2+, Mn2+, Mo, ca2+ etc. are very important, for they are either the integral part of some organic molecules or act as co-factor in enzymatic activities.
Certain organic factors, important for cellular metabolism, because many of the vitamins act as co-enzymes; these are required for enzymatic activities, without which cellular processes come to stand still.
Size and shape: The size of cells varies from 10 micron to many centimeters in length. For example, cotton fibers are several mm in length. The shape ranges from spherical, isodiametric, and hexagonal to tubular. This is genetically predetermined to perform different functions.
Cell structure:
When the cell is observed through light microscopes, which may have the maximum resolution of about 2000 times, very few details can be made out. On the other hand, if sections of the cells are observed through electron microscope, which has a resolution power ranging from 50,000 to 150,000 times enlargement, even smaller structures stand out clearly. Inspite of high resolution, it is not possible to make out all the structural details.
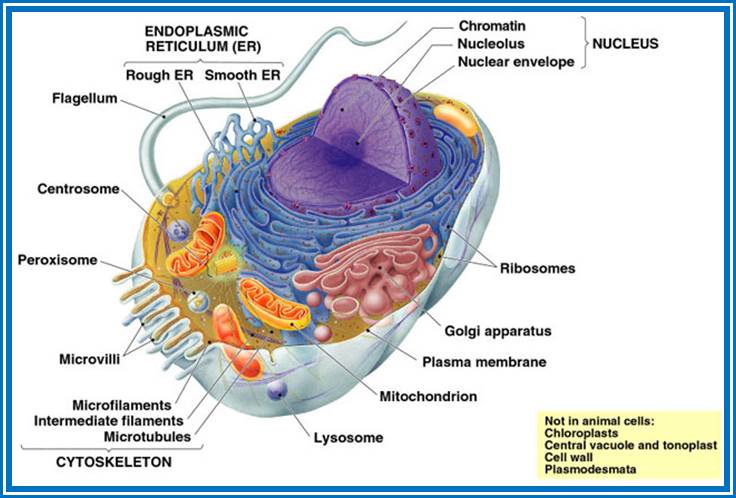
The following structures are found in the cell, 1) Cell wall, 2) Plasma membrane, 3) Nucleus, 4) Plastids, 5) Mitochondria, 6) Golgi complex, 7) Ribosomes, 8) Cytoskeleton, 9) Micro bodies, 10) Centrosomes,11) Endoplasmic reticulum, 12) Central Vacuole and 13) Non-living cell inclusions.
Cell wall: Only plant cells and bacterial cells posses a protective structure as the cell walls outside the plasma membrane. Bacterial cell wall is firmly adpressed to the underlying plasma membrane.
Its chemical composition and structure are more complex. However, it is basically made up of long polymers of glucosamine (NAG) and muramic acids (NAM), which in turn are cross linked by short-pentamer oligopeptides; thus, they form a mat like structural layers around the plasma membrane. Hundreds of such layers are deposited one above the other to form a very tough wall. Many bacterial cells produce a mucilaginous pectose layers outside the cell wall and this layer is called the capsule. But many bacterial cells do contain another lipid layer studded with proteins, oligosaccharides and glycol and proteoglycans.
![]()
But higher plants have cell walls mainly made up of cellulose fibers. Addition to these, pectin’s, hemicelluloses and lignin are also deposited on primary cellulose layer; the thickening is only at the later stages of development of the cell.
With proper staining, if a group of cells are observed, the cells appear to be held together by a kind of cementing material called middle lamella. It is made up if calcium
pectate. Next to calcium pectate layer, the cell wall that is laid later is differentiated into 2 or 3 distinct layers. They are primary wall, secondary wall and tertiary wall.
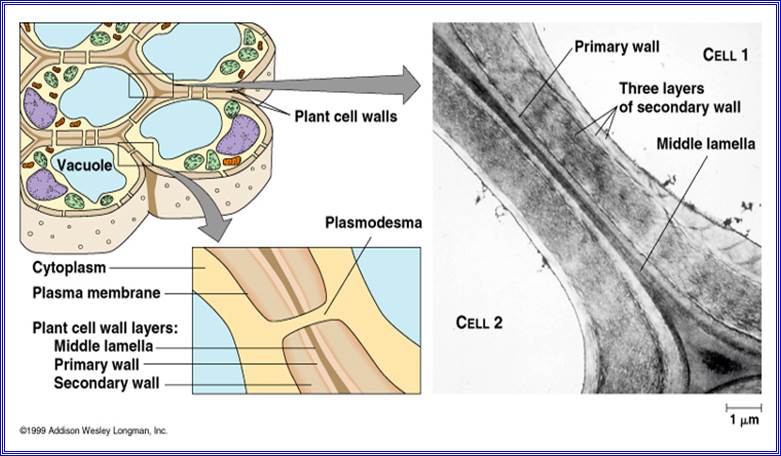
The primary wall is the first true cell wall to be deposited and it is purely made up of cellulose. With the development of the cell, the additional layers are deposited, called secondary and tertiary wall layers, like in the case xylem and sclerenchyma. Certain areas in the cell walls remain without thickening and these are called Pit areas, through which fine protoplasmic strands transverse across between two cells and provide a continuum to protoplasm; they are called plasmodesmata.
![]()
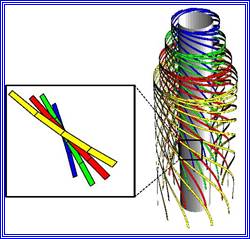
The primary cell wall, which is the first wall layer to be deposited, is made up of cellulose fibres. These fibres are considerably longer and they are deposited layers after layers; oriented longitudinally, transversely or obliquely. The deposition and the orientation of these layers are aided by microtubules that are found on the inner face of the plasma membrane.
Each cellulose fiber is made up of 8000 to 120,800 D- Glucose units, which are linked to each other by glycosidic bonds to form a long chain of glucose units, which show helical conformation. Hundreds of such cellulose threads are grouped into a bundle called micelles; these in turn are aggregated into micro fibrils which by further aggregation develop in to macro fibrils.
Such micro/macro fibrils are deposited regularly either longitudinally or transversely to form uniform layers. These fibres are embedded in a matrix made up of pectate substances and hemi-cellulose materials like poly-xylose and others.
Primary cell wall Secondary cell wall
Primarily cellulose Few layers of cellulose
Hemicelluloses Xylose, mannose
Pectic Complex lignins
Elastic Non-elastic
Laid on middle lamellae Laid over primary wall
1-3 micron thick >5-10 micron thick
Cell membranes:
These are the most important structures of the cell, for they are responsible for protecting and separating the protoplasm from the external environment.
It helps in selective uptake and transport of ions, provides surface area for many biochemical signal transduction reactions and also helps in specialized functions. In fact, most of the cell organelles are bounded by membranes. Notwithstanding this, a large number of functional molecules are integrated into these membranes.
Chemical composition:
Almost all membranes are basically made up of proteins and lipids. The ratio between proteins and lipids may vary in different cell membranes, but generally it is equal. It is not uncommon to see some carbohydrates as glycoproteins associated with membrane on the outer surface of plasma membranes. Proteins found in membrane are not of the same kind, but differ in their structure, chemical composition and function. Some of them are large and many of them may be smaller. However, most of the proteins are globular in nature having either hydrophilic or hydrophobic or both the characters. Lipids, on the other hand are of various kinds. The common lipids found in the membranes are phosphatidyl choline (Lecithin), phosphatidyl ethanolamine, Phospholipids glycerol, cholesterol, etc. some of the phospholipids exhibit hydrophilic groups at one end and hydrophobic at the other end. Nevertheless; the composition of lipids varies from membrane to membrane, for they have different functions to perform.
Membrane structure
The structural organization of various compounds with in the membrane was an enigma for a long time. With the advancement of biological techniques, it has been found that the membranes exhibit fluid-mosaic structure (Singer and Nicholson 1972). Basically, lipids form bilayers by organizing hydrophobic layers facing each other and the charged region outer surface. Proteins, depending upon the charged nature, some are found on the surface and some are embedded. But proteins & lipids of various kinds are oriented towards each other in such a way, they exhibit semi-solid (crystalline) and semi fluid properties. The arrangement of lipids and proteins is of mosaic pattern, where many proteins are half buried in the lipid bilayers and some traverse the entire cross section of the lipid layer in such a way a part it is buried in the core and some are located at the peripheral surface. The structural and chemical heterogeneity is the hall mark of these membrane structures. This model explains various biological phenomena observed in most of the biological systems.
Various types of membranes found in the cell, like plasma membrane, endoplasmic reticulum; organelle membranes exhibit basically the same structural pattern, but vary in their lipid and protein composition. Some of them are single unit membranes (plasma membrane, tonoplast, Lysosomes and peroxisomes) and some have double membrane systems (endoplasmic reticulum, nuclear membrane, golgi membranes chloroplast membrane and mitochondrial membrane). However, they show differences in their chemical composition structural features and functions.
Plasma membrane
This is the outer most membrane of the cell, within which all protoplasmic structures are included. In plants, cell wall acts as an additional protective layer on the outer surface of PM, but in animal cell, plasma membrane itself is the bounding membrane.
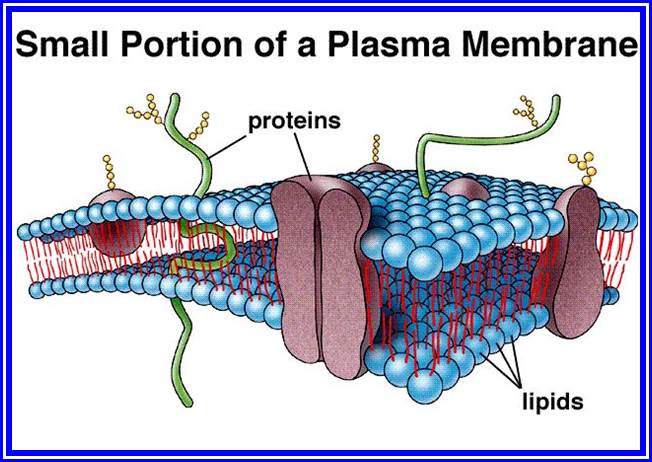
This membrane is on the inner surface of the cell wall and performs various functions like osmosis, selective absorption of various mineral nutrients, accommodates innumerable signal transducing receptors for various stimuli (electrical, light mechanical and chemical) and invariably exhibits dynamic properties. Proteins found in the plasma membrane are vectorial (directional) arranged. At some places, the plasma membrane shows inward projections and they are in continuity with the endoplasmic reticulum. Plasma membrane is also the site for pinocytosis and phagocytosis. Thus, the plasma membrane exhibits unique but varied properties of its own. It exhibits dynamic fluidity never to be constant and stagnant.
Endoplasmic Reticulum:
It is a labyrinthine net work of double membrane sheets. They are found in all living cells with the exceptions of mature erythrocytes and cells of bacteria. Endoplasmic reticulum (ER) occupies more than 50 to 90% of total cell volume. It is in contact with the outer plasma lemma and the outer nuclear membrane. ER is made up of two single unit membranes folded to form adpressed sheets, which enclose a channel or cisternae. ER is extensively branched, thus the surface area for reactions is enormously increased.

On the basis of presence or absence of ribosomes on the surface, two classes of ER can be recognized.
1) Smooth ER (SER) is without ribosomes, 2) Rough ER (RER) has innumerable ribosomes on its outer surface. These membranes are highly mobile and they undergo rapid flux, changing from SER to RER and RER to SER. Added to this, the entire ER exhibit continuous sweeping movements which help in the distribution of cellular components in the matrix. The ER membranes are supported by microtubule skeletal network. These membranes are highly dynamic and show rapid turnover.
Functionally ER exhibits various activities like synthesis of proteins, mechanical support for the fluid protoplasm, synthesis and storage of lipids, synthesis and storage and transport of proteins to different destination through golgi membranes. Some of the functions are detoxification, transport of various cellular components, formation of micro bodies, formation of secretory vesicles, cell plate and others. The above-mentioned functions indicate that the ER is at the heart of various cellular activities. Furthermore, during the development of nucleus and other cell organelles like, chloroplast, mitochondria, golgi complex, micro bodies, Lysosomes etc. ER provides membrane fractions to them. Exchange of membrane components among them is pervasive and common.
Golgi bodies
![]() A group of membrane cisternae, discovered by Camillo Golgi
in 1890, are called as Golgi bodies. They are present in all cells except
bacterial and blue-green algal
A group of membrane cisternae, discovered by Camillo Golgi
in 1890, are called as Golgi bodies. They are present in all cells except
bacterial and blue-green algal ![]() cells.
These structures vary in number from cell type to cell type. In secretory
tissues like thyroid and liver they are present in large numbers than in other
type of cells. They are abundant in secretory surfaces like stigma of the
pistils.
cells.
These structures vary in number from cell type to cell type. In secretory
tissues like thyroid and liver they are present in large numbers than in other
type of cells. They are abundant in secretory surfaces like stigma of the
pistils.

Electronic-Micrography
Golgi bodies are made up of a group of stacked membrane cisternae. Some times these membranes structures show extensive reticular network. Generally, the distal ends of double membranes are dilated into vesicles and some of the vesicles are in the process of pinching off. An interesting feature is that the Golgi complex is surrounded by ER and at some places they look like in continuity with ER, but these proximal membranes of ER are free from ribosomes called SER (smooth ER).
The stacked Golgi membranes have two faces, i.e., formation face is called cis face and maturation face as trans face. The formation face has convex surface and has a number of small vesicles pinched off from SER. In fact, certain proteins synthesized on the RER, enter into the lumen of ER and hence they are transported in the lumen towards the transitional ER which are in close association with Golgi complex and then the protein containing ER membranes pinch off as vesicles. These in turn fuse with one another or with golgi cis membranes and develop into membranous sacs. Within these membranous sacs, proteins and such products get further modified. Later such products are sorted out and get enclosed and pinched off in the form of vesicles from maturation face called Trans surface of the Golgi complex. Similarly, many secretary products that are synthesized on RER and transported into Golgi complex, later the matured products get enclosed in vesicles and budded off. The golgi derived vesicles are loaded with proteins that are specifically targeted to various destinations.
Thus, Golgi bodies perform various complex processes, like glycosylation of proteins, synthesis of cell wall polysaccharides, maturation of zymogen granules, formation of primary lysosomes, secretion of lipid bodies, acrosome formation, neural secretion etc. Nevertheless, the participation of ER is every essential in the function of Golgi membranes. Golgi membranes are associated with transport of proteins from one site to the other. In all the above-mentioned processes, packing, maturation and secretion of specific substances are the most important events of Golgi functions. The golgi complex is at the heart of membrane flow.
Lysosomes:
Though these organelles were noticed in 1949, later it was de Duve who coined the term Lysosomes for such dense bodies. Lysosomes are often called suicidal bags, misnomer, for they are capable of digesting various cellular structures and digest every food etc., if they are damaged and make to break open to release the contents.

Phagocytosis and Autophagy

Phagocytosis and Autophagy
Without any exception, all eukaryotic organisms contain these organelles in their cells. These are found in various sizes. Lysosomes are bounded by a single unit membrane and enclose a group of hydrolytic enzymes. Paradoxically, the surrounding membrane is not digested by the enclosed hydrolytic enzyme; this is probably due to special modifications of the membrane and lysosomal fluid which is more acidic. The intactness of the membranes is mainly dependent upon certain membrane stabilizers like cholesterol, cortisones, cortisols, vitamin E, antihistamines, heparin etc. On the other hand, substances like Vitamin A, Vitamin B, Vitamin K, B-estradiol, testosterone and digitonin labalize and cause leakiness. Sometimes at higher doses, the membrane may completely disperse and all the lysosomal contents may be released. As a result, the cell may be completely digested.
Lysosomes are important cell organelles in digesting various macromolecules like carbohydrates, proteins, fats, DNA, RNA and others. The breakdown of these molecules during various stages of development and metabolism is governed by the controlled release of these enzymes.
The origin and development of lysosome itself is controlled by many environmental factors. For example, when an organism is starved of food, lysosomal number increases tremendously. This is in order to degrade whatever food material that is available in the cell. Similarly, when yeast cells are subjected to anaerobic conditions or starved of food, within minutes, Lysosomes increase in number and actively; they chew up all the available materials including mitochondria. In the case of germinating castor seeds or maize grains the increased lysosomal activity helps in the degradation of fats and starch into simple molecules for the growing seedlings. The number of enzymes and the kinds of enzymes found in each of lysosomes is not same or constant. The contents vary depending upon the tissue and the metabolic status of that tissue. Commonly available lysosomal enzymes are nucleases, phosphotases, lipases, proteases, glycosidases and sulfatases. Even some of the condemned proteins are taken in through L.AmP proteins on its membrane surface and digest the same. Many of the lysosomal enzymes are released in a regulated way and they have definite pH optima for their peak activity. Lysosomes and some trans golgi vesicles and incoming endosomes form a kind of network which provide material for its growth and maintenance.
Curiously enough, lysosomes take their origin from Golgi bodies. Lysosomes enzymes are synthesized on RER: then they are transported to smooth transitional vesicles. Afterwards they are integrated into Golgi sacs at the formation face. After undergoing modifications and packing, they are then sorted into vesicles, which are budded off from maturation-face of the Golgi complex. The vesicles containing lysosomal enzyme are marked by the addition mannose6-phosphate, such vesicles ultimately dock with lysosomes via late endosomes or directly. These may further fuse with one another to form larger structures or they may fuse endosomes or and with phagocytotic vesicles (phagosomes) to form secondary lysosomes, where the engulfed substances are digested and the products are resorbed into cytoplasm. The undigested materials are removed by defecation. Many a times, the lysosomes move towards the plasma membrane and unload all their contents, thus they cause extracellular digestion. Lysosomes also play a significant role in the acrosome formation (cap structure) of spermatozoid. The presence of such cap structures in sperms help in their penetration through the tough cortical walls of the egg cells.
Lysosomes are also known to play a very important role in metamorphosis of amphibians and insects. For example, during the transformation of tadpole into adult frog; the long tail of the tadpole gets digested by the lysosomal activity, the process called resorption. Recent investigations have further shown, that the increased activity of lysosomes causes severe destruction of tissues, probably of lysosomes cause severe destruction of tissues, probable break down of chromosomes leading to abnormality; perhaps even cancer may be induced. Another instance of lysosome induced disease is Rheumatoid arthritis (Joint pain). Thus, lysosomes play a significant role in the metabolism and development of an organism. One can also find various lysosomal based human diseases.
Microbodies;
These are spherical, electron dense, granular bodies. They are bounded by a single unit membrane. Such structures are found in both animal and plant cells. Their number varies from 50 to 100 per cell; their number can increase or decrease based on the requirement. Germinating seed cells show maximum numbers; often they form a link between chloroplast and mitochondria, where peroxisomes are involved in the detoxification of oxygen free radicals by catalase or peroxidase activity.
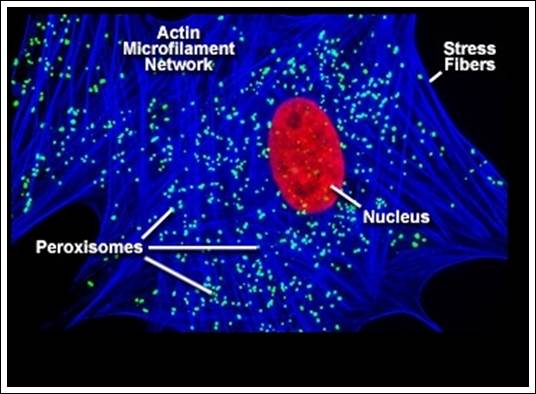
There are two types of micro bodies and they are characterized by their functions viz., 1) peroxisome, 2) glyoxysomes; the former exhibit peroxidase activity and the later show glyoxalate activity. Such bodies are found in various tissues like liver, kidney, intestine, brain, lung epithelial cell, testis, brain, adipose, and photosynthetic cells of green plants.
In C3 plant cells (Calvin plants) they are closely associated with mitochondria and chloroplasts. They are responsible for photorespiration. This process severely results in the depletion of photosynthetic products. However, they are not that active in C4 plants. Nevertheless, in C3 plants both Glyoxysomes and Peroxisome act co-operatively utilizing Ru-DP glyoxalate and breaking it down to glycollate, which is then metabolized by peroxisome. On the other hand, in animal tissues like liver and other cells, various substances like urea, amino acids, lactic acid etc. are oxidized by peroxisome to H2O2. In this process oxygen is utilized. As H2O2 as peroxide is fatal to living cells, it is salvaged or removed by superoxide dismutase (SOD) reaction and oxidation to water by utilizing substrates like ethyl alcohol, methyl alcohol, nitrates, etc. The presence of these structures and their function is fascinating, for their exact role is not clearly understood. They are also implicated in thermoregulation of certain organisms.
Ribosomes:
Ribosomes are ultramicroscopic cellular organelles, first observed by Palade. Though they are submicroscopic in size, they are extremely important, for they are responsible for the synthesis of proteins.
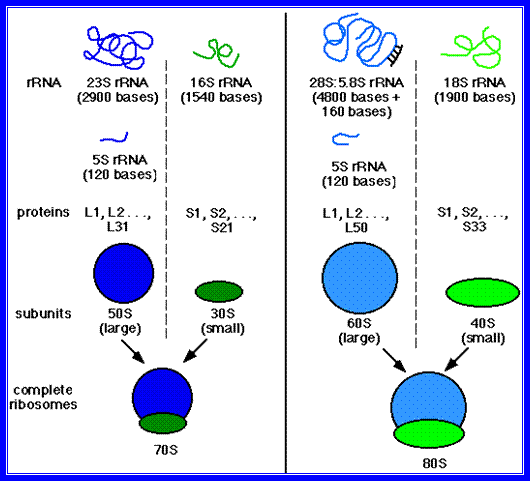
Ribosomes are found in millions, in eukaryotic cytoplasm, but in bacteria, ribosomal count is approx. 20000. They are also found in organelles like chloroplasts and mitochondria. These structures are either roughly spherical or ovoid in shape. They are very stable and can remain functional at least 120 days or so. Basing on the molecular weight (determined by equilibrium density ultra centrifugation) they have been broadly classified into two types, i.e., 80S and 70S types. The S-values of the slightly vary among the organelle ribosomes. Ribosomes of 80s type are exclusively found in eukaryotic organisms.

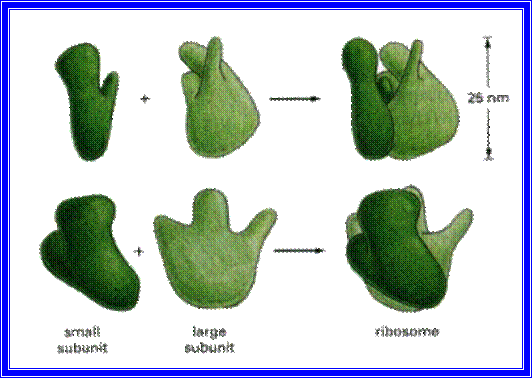
Structurally both 70S and 80S ribosome look alike.
But 70s ribosomes are restricted to prokaryotic organisms like bacteria and blue-green algae. Still smaller ribosomes are found in mitochondria and chloroplasts, but their sizes vary.
When functional ribosomes are subjected to Mg2+ depletion, they separate into large and small subunits. If the concentration of Mg2+ is increased the free subunits reassemble into functional units. The concentration of Mg2+, if further increased, ribosomes aggregate into dimmers, tetramers or octamers, thus one can precipitate ribosomes and collect them by centrifugation.
The smaller subunit under electron microscope exhibits a shape of elongated cucumber with an indentation and a twist. But the larger subunit shows a shape of a mother in sitting position with the knees upright, having the baby on her lap. Here the ‘Baby’ is equivalent to a smaller submit (note this purely my imaginary explanation not found in any text books). The larger subunit has a grove in the middle through which the nascent polypeptide traverses and exits.
Ribosomes found in the cytoplasm exist either in membrane bound state or free state. The ratio between these varies from cell to cell type and depends upon the functional state of the cell. But in bacteria most of the ribosomes exist in free state.
Chemical analysis of these structures indicates that they are made up of proteins and ribose nucleic acids (rRNA) roughly in equal ratios. The proteins of larger subunit of prokaryotic bacteria consist of 34 subunits called L1 to L34 and the smaller subunit has 21 factions, s1 to s21. Each one of these proteins is unique with the exception of two proteins i.e., S 20 and S 26 which are present in both the ribosomal units.
Ribosomal RNA exists in three or four different sizes which are expressed in Svedberg units (S) as shown in the table below.
The RNAs present in ribosomes show various sizes, and most of them are rich in Guanosine and Cytosine nucleotides. In prokaryotes rRNAs coded for by seven operons as precursor containing 16s, 23s and 5s RNA. Such precursors also contain tRNA blocks tucked in them. The genes responsible for the synthesis in eukaryotic cells are found in multiple copies ie. of 200-500 organized in tandem repeats. Furthermore, rRNA genes are exclusively found in secondary constrictions or nucleolar organizer region of the chromosomes, in humans they are located on chromosomes 13, 14, 15 21 and 22. The rRNA genes contain 28S RNA, 5.8sRNA and 28sRNA gene, but the 5 S RNA genes are found elsewhere and spread over in other chromosomes. These RNAs are transcribed on rRNA genes as larger precursor molecules (45S RNA) and then they are spliced and processed into smaller molecules such as 28 S, 18 S and 5.8 S RNAs. Notwithstanding this, the ribosomal RNA genes in the case of Frog oocytes are amplified 1000-fold. Enormous number of ribosomes are synthesized and accumulated in the eggs. Another interesting aspect of ribosomes is that, various ribosomes proteins are coded for by the chromosomal DNA other than the nucleolar DNA. They are synthesized in cytoplasm and they are transported into nucleolar region of the nucleus. There, the rRNA and ribosomal proteins associate to form functional subunits of ribosomes in a hierarchical fashion. Later they are transported to cytoplasm through nuclear pore complexes to perform protein synthesis.
However, mitochondria and chloroplast employ a different mechanism for the assembly of ribosomal components. Organelles synthesize their own rRNAs and get imported riboproteins from cytoplasm and assemble functional ribosomes which more or less similar to prokaryotic ribosomes in size and function. Their activity can be inhibited by chloramphenicol.
The quintessential function of ribosomes is to act as dynamic machinery for the synthesis of proteins. This is achieved by the association of ribosomes with messenger RNA (m.RNA) and amino acid loaded transfer RNAs (aat.RNA). Many ribosomes may be associated with a single m.RNA and such a cluster is called polysome. These polysomes may be free from membrane, or membrane bound.
Plastids:
Plastids are very important cell organelles found mostly in plant cells. They are mainly responsible for photosynthesis.
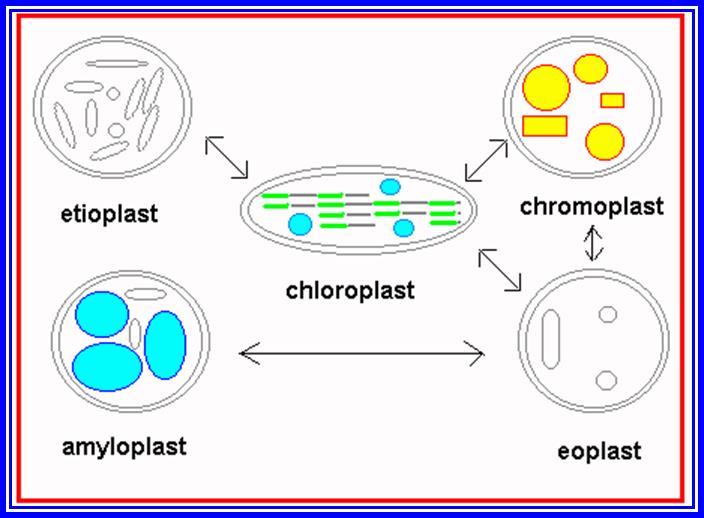
Proplastids to different Plastid’s forms.
Green color of plants is due to the presence of green pigments in plastids. Other colors found in various structures like leaf and flowers are due to the presence of other colored pigments in the plastids (other than green). Basing on the presence or absence of pigments, plastids have been broadly classified into colorless plastids (Leucoplasts) and color plastids (Chromoplasts). If the plant tissue containing Leucoplasts are exposed to sunlight, they may turn into chromoplasts. On the contrary, if plants with chromoplasts are kept in dark for sufficient time, the chromoplasts will be transformed into colorless plastids.
Leucoplasts are generally found in roots and inner tissues, which do not receive sunlight. Such plastids normally store different kinds of food materials like starch, proteins and oils; basing on this they are called Amyloplasts (store starch), Proteinoplasts (proteins) and Elaioplasts (oil). Such structures are also found in abundance in various storage organs like fruits, tubers and oil seeds.
Chromoplasts, on the other hand depending upon the dominant pigments present, are classified into chloroplasts (green) Rhodoplasts (Red), Fucoplasts (brown), Xanthoplasts (Yellow) and so on. Majority of terrestrial plants and aquatic plants like green algae contain chloroplasts. It is important to note that plants found in ocean which account for the major part of vegetation on this planet, contain different kinds of chromoplasts and also green plastids. While eukaryotic plants (higher plants) contain well developed plastids, plants like photosynthetic bacteria and blue green algae (prokaryotic) do not possess such plastids, but still they perform photosynthetic functions because the photosynthetic pigments are either found organized in the form of stacks of membranes or as vesicular structures called chromatophores.
Number: The number of plastids, particularly in green plants, varies from one to hundred or more. Plants like Chlamydomonas have only one chloroplast; spirogyra has two spirally coiled chloroplasts and in higher green plants like angiosperms the number varies from 20 to 100 per cell.
Shape: Chloroplasts show some interesting variation in their shape. This shape is constant for a given species. For example in Chlamydomonas, it is cup shaped; in spirogyra it is spirally coiled and ribbon shaped, in Zygnema it is star shaped, in Hydrodictyon it is reticulate type and in higher plants it is oval or spherical. Shape and size of chloroplasts is species specific.
Size: It varies from 0.5 micron to 6 micron in diameter but again its fully formed has specific sizes and species specific.
Structure: Basically, most of the chloroplasts are bounded by two single unit membranes and the space between them is called periplastid space. Inside the membrane system, chloroplasts are filled with a highly complex fluid called stromatal matrix or simply called stroma. Within this fluid, various specialized structures, macro molecules and enzymes are either suspended state or the fluid shows flux. The structures found are grana, ribosomes, DNA, RNAs, starch and other various enzymes, coenzymes required for carbohydrate and fatty acid synthesis and amino acids.
Grana: These are the most specialized membrane systems derived from the inner chloroplast membrane. A single chloroplast may contain 10-30 grana. Each granum is made up of 10-60 circular membranous discs stacked one above the other and such discs are called thylakoids. The grana are inter-connected by the thylakoid membranous extensions called inter-granal lamellae or stromal lamellae. However, in C4 plants, like sorghum, sugarcane and other tropical grass plants, chloroplasts show two types of organizations, i.e., the chloroplasts found in the bundle sheath cells do not possess any differentiated granal structures, but are filled with diffused stromal membranes, arranged parallel. However, chloroplasts found in mesophyll cells show granal structures. Chloroplasts in bundle sheath show centrifugal arrangement, i.e., chloroplasts are oriented towards the mesophyll cells. Protoplasmic connection is found between mesophyll cells and bundle sheath cells.
Thylakoid: Thylakoids are the highly differentiated and say, super specialized circular membranous cisternae, in which various molecular complexes are embedded and they are responsible for photochemical reactions. Though the membrane has the same basic components and organization as that of any other unit membrane, it is the assembly of photosynthetic pigments and its associated proteins makes it a specialized membrane system. But photosynthetic pigments like, Chlorophyll-a Chlorophyll-b, Carotenoids and their associated proteins are grouped in such a way that they act as units for performing photosynthetic functions. Based on the chemical composition, size, structure and function, they are classified into Photosystem I and Photosystem II. Each of these photosynthetic units contain about 250-300 chlorophyll molecules associated with specific and specialized proteins. These photosystems appear to be granular in structure of different dimensions.
If thylakoid membranes are cut open by freeze-fracture method and observed under electron microscope they reveal the presence of two types of photosynthetic units of different dimensions and they are called Quantasomes The larger quantasome, which is found on the inner surface has a size of 185A^o. They are grouped into an array of 4 to 6 units. On the other hand, the smaller particles are of 110 A0 size and are found arranged on the outer surface of the membrane. The former particle has been identified as photo system II and the later as photo system I. The outer surface of the thylakoid membrane i.e. surface A is also studded with granular particles of different sizes. They have been identified as RuBP carboxylase and ATP synthetase units. However, PS I and PS II are arranged in such a way they fit into one another like Jig-saw structures. This ‘close and tight’ fit arrangement helps in the co-coordinated functioning of the photosystems. But the inter-granal lamella contain only PS I. The chemical composition of PS I and PS II and their function is discussed in greater detail in the chapter photosynthesis. There is another important complex called Cytochrome b6-f complex which acts between these two systems and it is found both in stromal and thylakoid membranes.
Plastid Nucleic Acids: Apart from granal structures, various types of nucleic are found in the stroma. The presence of a circular double helix DNA molecule of 123 to 200Kbp long is highly significant. This gives the chloroplasts a status of semi autonomy. Such 10 to 30 circular DNA molecules are found in each chloroplast. It has been estimated that the information present in DNA is sufficient to code for ~120 plastid proteins or more. The relationship between certain segments of DNA and certain proteins of chloroplasts has been established, however the knowledge about the other regions is not known. Majority of RNAs like r.RNA, tRNA about 120 species of m.RNA, are coded by chloroplast DNA. The presence of various types of RNAs and 70s ribosomes, clearly indicates that chloroplasts are endowed with a machinery, which can synthesize most of its proteins by itself. Nonetheless, majority of the proteins that are required for chloroplasts to function are synthesized on cytoplasmic 80S ribosomes and transported into plastids via chloroplast membrane transport systems. The interaction and regulation of gene activity between nuclear genes and plastogenome is an upcoming field of molecular biology, whose scope is unlimited.

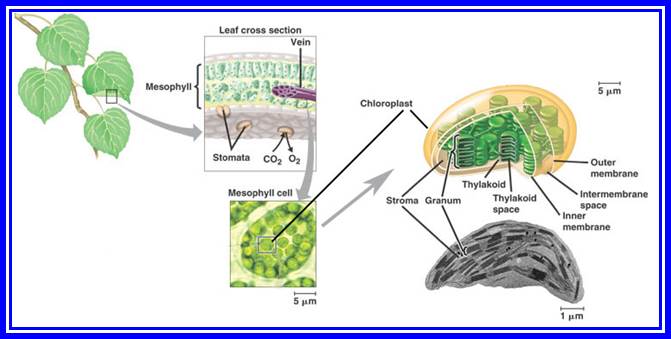
Structural features of Plastids
Apart from the above said structures and molecules, the stromatic fluid also contains an army of enzymes responsible for carbon pathway, amino acid synthesis, starch synthesis, protein synthesis, fatty acid synthesis and other enzymes for nucleic acid metabolism. Among them the RuBP carboxylase is found in very large amounts. In fact, it has been considered as the most abundant protein in nature, next to only to Tubulins. This RuBP enzyme protein accounts for more than 50% of the total leaf proteins.

This figure summarizes Light reactions:
Biogenesis: Various plastids, during biogenesis, develop from the pre-existing plastids called proplastid. They are, in fact, inherited from the maternal side. While the meristematic cells divide, proplastids too, divide by binary fission, just like cyano-bacteria, and get distributed equally or unequally to their daughter cells. When tissues or cells containing such proplastids are exposed to sunlight, particularly blue and red lights, they undergo a series of developmental changes. Proplastids contain a double membrane system. With the onset of light, the plastogenome gets activated; later certain nuclear genes also get activated required for plastid development. The future of it depends upon the kind of tissue in which it is located and other factors, where a plastid can develop in to colored, other than green, into colored plastids, as found in flowers. Activation transcriptions not only in the plastid, but also in nuclear genes. This leads to the transcription and translation required for plastid biogenesis. The plastid membranes have protein transport system located in a region where one finds the membranes in close association called attachment points. It is here cytosol protein transporters are located. It is at this time one can observe the inner proplastid membrane starts producing inward invaginations in the form of finger shaped processes. They are loaded with imported proteins. These invagination in turn start pinching of membranous vesicles in large numbers. These vesicles then start expanding and later arrange into stacks. Finally, they get organized into granal and intergranal structures. Along with these developments, various stromatal ingredients are also synthesized and accumulated. The regulated interaction of nuclear as well as plastid gene activity is a must for the development of functional chloroplasts. Chloroplasts perform light reactions where light energy captured, converted into chemical energy and stored in energy rich bonds of ATP and reducing power NADPH+H. Besides photosynthesis, they also perform carbohydrate synthesis and fatty acid synthesis and transport.
Mitochondria
Most of the biological activities are energy dependent and the energy that is required is the chemical free energy. The provision and production of such energy rich molecules like ATP, PEP, PGALD, Succinyl Co.A., Acetyl Co.A, NADH2 and such molecules are must for the sustenance of any cellular function.

Mitochondrial membranes, Outer and Inner; the inner has inward extensions loaded with functional proteins.
Mitochondria are one of the most important cell organelles, which are endowed with a capacity to produce energy rich molecules like ATP and NADH+H. In fact, mitochondria are considered as ‘power plants’ of the cells. Besides producing energy rich molecules, it also performs fatty acid oxidation to release energy rich components and other intermediate molecules which are essential for other metabolic pathways.

Furthermore, mitochondria are also involved the synthesis of amino acids.
Number, size and shape: The number of mitochondria per cell ranges from few to many hundreds or more and depends upon the type of the cell and its metabolic status. Whenever or wherever, there is a need for higher amount of energy output, mitochondria are found in large number, example: flight muscles; 1000 or more per cell. On the other hand, liver cells contain little less number. Similarly, mesophyll cells of plants, where greater number of chloroplasts are present, the mitochondria are in negligible numbers, for chloroplasts themselves satisfy most of the energy needs of the cells. Similarly in anaerobic yeast cells there are few mitochondria.
The size of mitochondria is generally between 3 –4 x 0.5 – 10 microns, but under certain pathological conditions the mitochondria may enlarge considerably, example: diseased thyroid glands. Normally mitochondria under optical microscopes with a resolution of 200 to 2000 times, appear to be thread like, similar to that of bacilli bacteria, provided they are stained with specific stains. But other shapes like spherical, branched and circular are not uncommon. Though mitochondria are stable, very often they exhibit a rapid turn over, involving changes in the size, shape and number.
Structure: Mitochondria are bounded by two single unit membranes. The outer membrane is smooth, but the outer surface may be studded with some granular structure, probably involved in glycolytic steps of respiration. The space found in between the outer and inner membranes is called peri-mitochondrial space. It is filled with a fluid and rich in cyt.C. The inner membrane is highly folded inwards called cristae. The number of cristae is not constant and it depends upon the demand for the energy supply. Nevertheless, the increase in cristae increases the surface area of reaction within a limited space and volume, an efficient system indeed.
Cristae: The inner membrane or cristae is made up of lipids and protein structures. Some of the lipids present- are phosphatidylinositol, phosphatidyl-choline, phosphatidylinositol and cardiolipin. With regard to proteins, the entire electron transport chain enzymes and its associated oxidative phosphorylation enzymes like ATP synthetase, coupling proteins etc., are assembled in this membrane. Particularly the ATPase enzyme which has a globular head and a basal stalk are found arranged uniformly on the inner surface of the inner membrane. These structures were once called elementary particles, but now they are referred to as Racker’s Particles (named after the discoverer). The interaction and regulation of gene activity between nuclear genes and mitochondrial genes is an upcoming field of molecular biology, whose scope is unlimited.
Electron Transport Systems: The proteins associated with oxidative phosphorylation chain are grouped into four complexes which are arranged sequentially.
Complex I and ComplexII--> complex III--> complexIV/V:
There are five such complexes: Complex I: It is made up of NADH + H reductase containing iron-sulphur (Fe-S) proteins and a flavo-protein with FMN as its prosthetic group. Complex II: It is made up of succinate Q-reductase with FAD and Fe-S proteins. Complex III-: It consists of cytochrome b, cytochrome c, and two Cu2 ions. ComplexIV: It is made up of cyt.a and cyt.a3 oxidases, Complex V: This is a complex structure made up of globular head piece, a stalk piece and a basal plate. The head piece, which is found as a globular structure projecting inwards from the inner surface of the membrane into mitochondrial matrix side, and it is made up of multiple protein subunits. When this is associated with the membrane, it acts as ATP synthetase, but if it is freed of membrane, it acts as ATPase (splitting ATP molecules). The basal plate is buried in the membrane and consists of hydrophobic proteolipids and contains proton transporting /secreting proteins. The head and basal plate is connected by a small stalk like structure called oligomycin-sensitivity conferring protein (OSCP).

In this chain of respiratory components, enzyme Q(complex) is found to act as a shuttle molecule between complex I and III; and complex II and complex III.
Similarly, another free and mobile molecule that is found between complex III and complex IV is Cytochrome C. Topographically these components are arranged vectorially. One can observe the sidedness of H+ proton release.
Mitochondrial matrix: The mitochondrial chamber is filled with a fluid called mitochondrial matrix. Inspite of the presence of numerous cristae, the mitochondrial chamber is not compartmentalized, and the matrix is continuous within the mitochondrion. The matrix is rich in proteins, because all the enzymes responsible for Kreb’s cycle are located in it. In addition, other enzymes responsible for amino acid synthesis, fatty acid oxidation, and DNA, RNA and protein metabolism are also found.
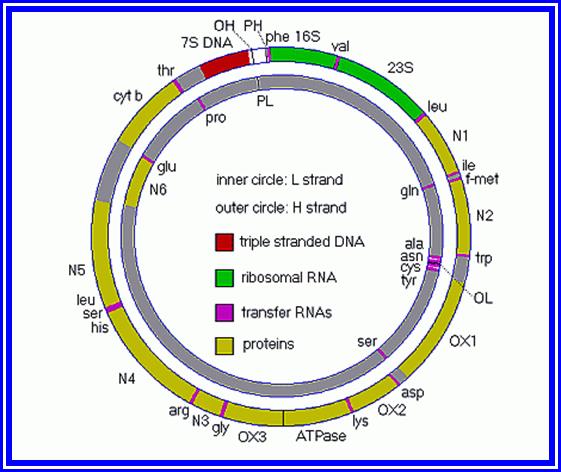
Diagrammatic Representation of Mitochondrial Genome with genes:
Other significant components of mitochondrial matrix are a circular DNA double helix, tRNAs, mRNAs and 70s ribosomes. The presence of the genetic material of 65KBp long to 200Kbp or more, depending upon the species provides the required components of proteins supplementing the input from nuclear genes; give the mitochondria a semi-autonomous status. However, the nuclear genome products are required in greater numbers and types for the complete development and function of mitochondria. Hence, mitochondria are considered as semi-autonomous organelles similar to that of plastids.
Origin and Biogenesis of Mitochondria:
Earlier views about the origin of mitochondria from the plasma membranes about nuclear membranes are now discarded, for there is neither substance nor facts to substantiate such claims.
Mitochondria in Divisional mode.
Recent studies have unequivocally showed that mitochondria take their origin in the cells from either pre-existing mitochondria or promitochondria. During mitochondrial biogenesis, genes of both nuclear genome and mitochondrial genome products interact. Though mitochondrial DNA codes for some 13 mitochondrial proteins, tRNA & rRNA, other components are coded for by nuclear genes, and they are synthesized in cytoplasm. Then they are transported into mitochondria through mitochondrial membrane transport complex, to form mitochondrial functional structures.
Cell Vacuoles:
Vacuoles are the spaces within the cells surrounded by single unit membranes. Don’t consider them as empty spaces. In plants, mature cells have a single large centrally located vacuole occupies nearly 50-70% of the cellular volume. It is filled with cell sap. It is separated from the rest of the cytoplasm by a single unit membrane called tonoplast.
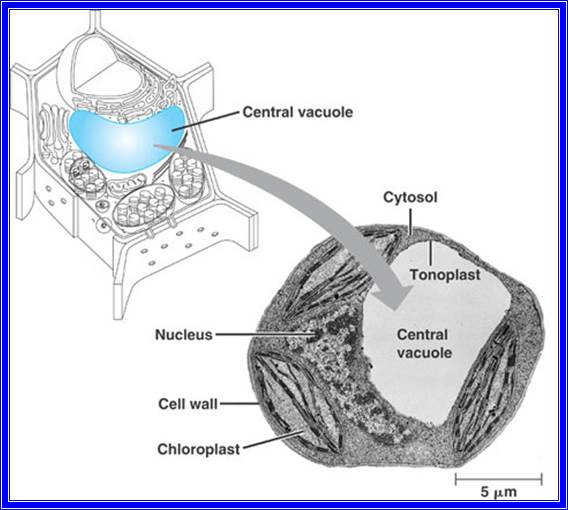
Tonoplast- central Vacuole
In meristematic cells, vacuoles are absent to being with,
but as the cell expands, a number of small vacuoles arise from the endoplasmic reticulum; later as the cells further enlarge, smaller vacuoles fuse with one another to form a single large vacuole. The content of the cell vacuole is important in maintaining the turgidity of the cells for they store water and its soluble forms. Furthermore, these vacuoles have been found to show lysosomal activities too. Nevertheless, vacuoles found in animal cells have drawn the attention, because they perform some important functions. Contractile vacuoles and food vacuoles are notable among them. Contractile vacuoles are helpful in the excretion and others help in digesting the ingested food materials. Thus, vacuoles play different roles in different organisms.
Cytoskeleton:
Microfilaments and Microtubules: Organisms, whether they are plants or animals, are made up of various types of cells, which have various shapes and functions. But all these cells are derived from the same mother cell called zygotic cell. The shapes and functions of these cells is defined and determined at the time of differentiation (the topic is more complex). Within these cells, cytoplasm with its components like Intermediate filaments (IF), microtubules (MT and microfilaments (MF) play important roles in determining cellular shape, structure and function. These structures act as the internal cytoskeleton and give a kind of mechanical strength to the cytoplasm and provided directions for the movement of cytoplasm, which makes the cytoplasm as a dynamic fluid.
Microtubules: Microtubules are fine tubular protein structures having dimensions of 200 A0 in diameter and many microns in length. The wall of the individual tubular structure is made up of 13 protein subunits called tubulins which exist in two forms called µ and b subunits; they are arranged alternatively. Hence the basic building blocks of microtubules are Tubulins of molecular weight 55000-58000 daltons. These subunits alternatively get polymerized at the membrane bound nucleating centers into tubular structures. This polymerization can be inhibited by colchicine, an alkaloid extracted from the tubers of Colchicum autumnale.
Such microtubules are dispersed and distributed in different patterns in the cytoplasm. Some form a kind of network in the cytoplasm, some are oriented towards nuclear membrane, and some run parallel to the plasma membrane in longitudinal direction of the cell. These structures being quite rigid, they are able to provide a structural support to various cells. Microtubule are associated with Endoplasmic reticulum for its sustainability and structural stability, so also Golgi membranes.
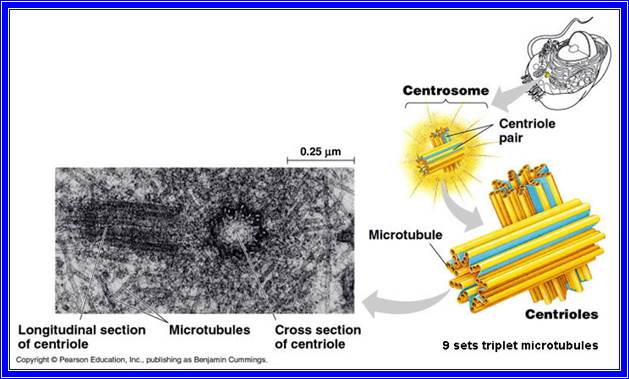
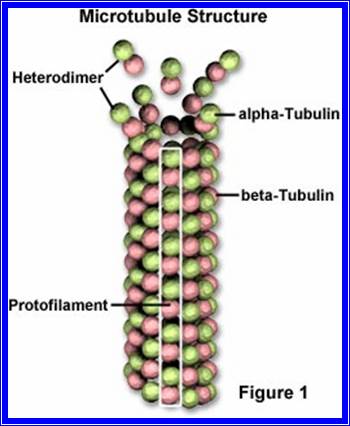
Microtubular structure
Notwithstanding being the cytoskeleton structures, they are also involved in the structural organization of cilia, flagella, centrioles, basal granules, mitotic apparatus, neurotubules etc. Such being the case, the function of not just being cytoskeleton support to the aqueous cytoplasm, but they are also involved in various functions like maintenance of the cell shape, cytoplasmic fluidity, membrane movement, chromosomal movement, cytokinesis, cell wall deposition, flagellar movement, sensory transduction, transport of cellular materials, cell polarity, morphogenesis, cell movement and others to say the cytosolic goods transported to the specific destinations and they aided by motor proteins such as kinesins, dyeinins and few more. They are involved in transport of goods along the MTs to their respective destinations. Tubulins are the most abundant proteins in the organic world next to RuBP carboxylase.
Microfilaments: Along with microtubules, cytoplasm also contains category of thin protein fibres called microfilaments. They represent the contractile system of the cytoplasm. Microfilaments are very fine protein filaments of 6-10 nm thickness and 10-100nm in length. These are made up of actin subunits and they can be associated with myosin and troponin proteins. Actin accounts for more than 20% of the total cellular proteins. Actin consists of basic building blocks called globular proteins called G-actin, of molecular weight 45 Kd. These globular units, by undergoing polymerization, develop into fibrous actin called F-actin. Normally two such F-actin proteins are helically coiled to each other and such filaments are associated with troponin. These filaments are further decorated with myosin proteins with calcium binding sites and ATPase proteins.
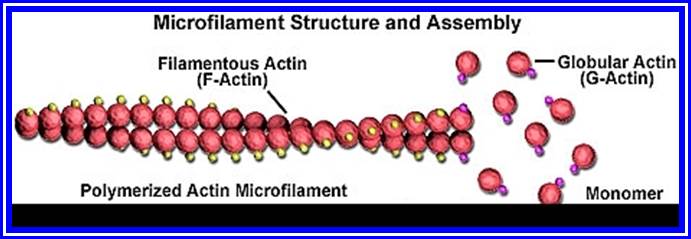
Microfilaments- contain Globular Actins organize into filaments.
These proteins are not just restricted to muscles, but they are also found in cytoplasm of almost all kinds of cells. The interaction between myosin and actin proteins generates the force for movement. The transformation of G-Actin to F-Actin and vice versa represents the interconversion of sol-gel system of the cytoplasm is the dynamic force for cytoplasmic fluidity.
The above said microfilaments are often located just beneath the plasma membranes in the form of bundles. These in turn are interconnected by a network of similar filaments which pervade the entire cytoplasm. These are also in contact with small membranous vesicles, microtubules, nucleus, polysomes, root lets of flagella etc. However, mitochondria appear to be free from such filaments.
These microfilaments, particularly actin filaments undergo depolymerization in the presence of an alkaloid called cytochalasin B. If this drug is supplied to living cells, it inhibits many cellular activities like cytoplasmic streaming, migration of cells, endocytosis, exocytosis and even cell-polarity fixation. The above said features clearly suggest the involvement of microfilament is vital in various cellular activities. For example, protoplasmic streaming in many plasmodia, plant cells and amoeboid movement of many protozoans is controlled by the activity of these microfilaments. Recent investigations have shown that in cancer cells the microfilaments are in a disorganized state, which certainly indicates the involvement of microfilaments the induction of cancer.
Centrioles: Centrioles are the characteristic organelles of animal cells. Invariably they are present in all animal cells, but they are almost absent in plant cells with the exception of some lower unicellular algae. They are found in the cytoplasm at one pole of the nucleus. It is made up of a pair of dark granular structures called centrioles, which in turn are surrounded by an amorphous region called centrosphere. Each centriole is made up of two open barrel shaped structure measuring 0.2 um m 0.5 um, sometimes they may be as long as 2um. These two structures are oriented at right angle to each other.
Structure: The wall of the open barrel shaped structure is made up of nine groups of microtubules arranged in a circle. Each group has three microtubules and skewed arrangement, towards the centre. These three microtubules are named A, B & C starting from the centre towards periphery. Compared to the proximal structure of the flagella, the central pair of tubules are absent. However, in the case of centrioles, which develop into cilia, the wall is made up of 9 groups of tubules and each has two microtubules and there are two centrally located microtubules. Chemically centrioles are made up of proteins like tubulin, dynein (ATPase) and other contractile proteins. Added to this, the report about the presence of DNA is very interesting. However, the chemical nature of the peripheral amorphous region is not known.
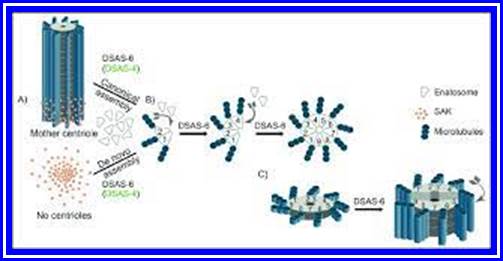

Research GATE;
Self-assembly and modularity in centriole assembly. Centrioles are barrel-shaped cylindrical structures buildup of nine triplets of microtubules. (A) SAK/PLK4 leads to canonical centrosome formation when a centriole is present (top image). In the absence of a centriole and in the presence of SAK/PLK4 activity, centrioles are formed de novo (lower image), pointing to the role of self-assembly in centriole biogenesis. (B) SAK/PLK4 recruits DSAS-6 which brings together the nine pre-centriolar units, enatosomes, organizing them laterally and longitudinally and specifying a tube-like centriole-precursor. These structures can recruit centriole microtubules perhaps through the action of DSAS-4 as suggested in C. elegans 7 (green). (C) Centriole elongation occurs through the addition of more enatosomes units (1', 2', 3', 4'), one by one, on top of the previous disc. Left image in (C) corresponds to a different perspective of a recently made centriole disc. https://www.researchgate.net/figure


Centriole propagation via duplication of preexisting centrioles
https://www.researchgate.net/

The dual role of centrioles in animals, either involved in cell division as part of the centrosome or in cell motility as part of the basal body of an axoneme. The figure on the left shows the structure of the centrosome consisting of two centrioles surrounded by the pericentriolar material (PCM). Each of the two centrioles consists of a ring of nine microtubule triplets, as shown in the insert in the middle section of the figure. The figure on the right shows how a centriole can attach to the cell membrane to act as a basal body and seed the growth of cilia and flagella. The microtubule skeleton within a cilium or flagellum is called the axoneme and has a similar nine-fold structure as the centriole, but with 9 doublets instead of triplets. Reproduced from Bettencourt-Dias (2013) with kind permission from the copyright holder. https://www.researchgate.net/
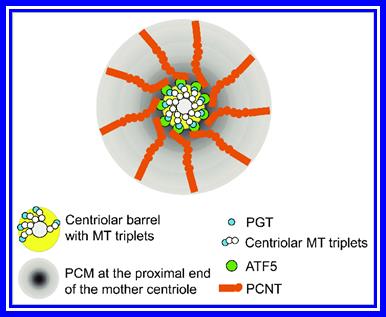
Simultaneous interactions between ATF5 and PCNT and between ATF5 and PGT promote PCM accumulation at the proximal end of the mother centriole. https://www.researchgate.net/
Centrioles have dual functions viz. 1) Cilia and flagella in lower organisms and other cells take their origin from centrioles. 2) During cell division, centrioles help in the organization of mitotic apparatus in a particular plane.
During cell division, particularly in animal cells, centrioles undergo duplication, so that centrioles produce procentrioles which are oriented at right angles to the mother centrioles. The organization of microtubules during the formation of daughter centrioles is not clear. Whether the message for the synthesis of these proteins resides in the DNA found in centrioles or in the nucleus is not clear. How the peripheral cytoplasmic ribosomes are involved in the synthesis and secretion of these proteins is again a mystery. Nevertheless, during cell division the two centrioles separate, at the same time a number of microtubular structures emerge from each of the centrioles and radiate in all directions and inter link with the two centrioles. Thus, these centrioles move away from one another towards pre-determined polar directions. However, the mitotic fibres that develop during cell division do not take their origin from the centrioles.
David Liu; https://www.ncbi.nlm.nih.gov/
Microtrabaculae:
David Liu
Micro trabecular structure of the axoplasmic matrix: visualization of cross-linking structures and their distribution;
Exoplasmic transport is a dramatic example of cytoplasmic motility. Constituents of axoplasm migrate as far as 400 mm/d or at approximately 5 micron/s. Thin-section studies have identified the major morphological elements within the axoplasm as being microtubules, neurofilaments (100-A filaments), an interconnected and elongated varicose component of smooth endoplasmic reticulum (SER), more dilated and vesicular organelles resembling portions of SER, multivesicular bodies, mitochondria, and, finally, a matrix of ground substance in which the tubules, filaments, and vesicles are suspended. In the ordinary thin-section image, the ground substance is comprised of wispy fragments which, in not being noticeably tied together, do not give the impression of representing more than a condensation of what might be a homogeneous solution of proteins. With the high-voltage microscope on thick (0.5-micron) sections, we have noticed, however, that the so- called wispy fragments are part of a three-dimensional lattice. We contend that this lattice is not an artifact of aldehyde fixation, and our contention is supported by its visibility after rapid-freezing and freeze-substitution. This lattice or micro-trabecular matrix of axoplasm was found to consist of an organized system of cross-bridges between microtubules, neurofilaments, cisternae of the SER, and the plasma membrane. We propose that formation and deformation of this system are involved in rapid axonal transport. To facilitate electron microscope visualization of the trabecular connections between elements of axoplasm, the following three techniques were used: first, the addition of tannic acid to the primary fixative, OsO4 post-fixation, then en-bloc staining in uranyl acetate for conventional transmission electron microscope (TEM); second, embedding tissue in polyethylene glycol for thin sectioning, dissolving out the embedding medium from the sections and blocks, critical-point-drying (J. J. Wolosewick, 1980, J. Cell Biol., 86:675-681.), and then observing the matrix-free sections with TEM or the blocks with a scanning electron microscope; and third, rapid freezing of fixed tissue followed by freeze-etching and rotary- shadowing with replicas observed by TEM. All of these procedures yielded images of cross-linking elements between neurofilaments and organelles of the axoplasm. These improvements in visualization should enable us to examine the distribution of trabecular links on motile axonal organelles. https://www.ncbi.nlm.nih.gov/
Intermediate filaments:
They are the most solid structural proteins involved in cellular cytoskeleton structural components as mechanical support system. Such elements in plants are more or less restricted to nuclear lamins located at the inner surface of the nuclear membrane.
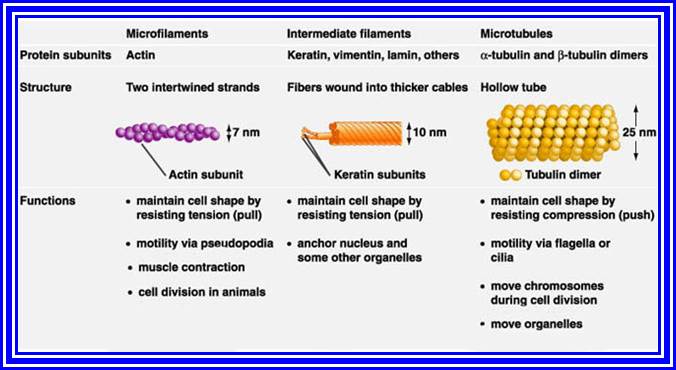
The centrosome is the major microtubule (MT)-organizing center (MTOC) in animal cells. It comprises a pair of centrioles and the surrounding pericentriolar materials (PCM), and is duplicated once per cell cycle in a highly spatiotemporally regulated manner. Because of the intricate links between Centrosome functions, mitotic spindle assembly and cell division, defects in the centrosome structure and function often lead to malformation of mitotic spindles and genomic instabilities, which result in a range of human diseases including tumorigenesis, ciliopathy, microcephaly, and dwarfism.1 Previous studies indicated that the proximal end of the mother centriole plays instructive roles in the biogenesis of the PCM and the procentriole and that the PCM is essential for centriole stability. However, the molecules and the mechanism that control centriole-PCM interaction are poorly understood. In our recent study, published in the current issue of Cell,2 we provided evidence showing that the activating transcription factor 5 (ATF5) acts unexpectedly as an essential structural PCM protein that bridges the PCM and the proximal end of the mother centriole. By interacting with both pericentrin (PCNT) in the PCM and the polyglutamylated tubulin (PGT) at the proximal end of the mother centriole, ATF5 controls the formation of the PCM and the integrity of the centrioles in a cell cycle- and centriole age-dependent manner. (Fig.at top 1). By David Liu
Microtrabaculae: High voltage electron microscope is a new version of electron microscope which has been now employed in the studies of cellular structures. There are few such microscopes that too only in U.S.A and each of them cost many million dollars. These microscopes are 32 feet in height, 20 tons in weight and produce million-volt electric discharges. The resolution power of the microscope is very high. A structure of 0.10-20 nm thickness can be made out clearly. This is a very useful technical innovation in the field of biology where a whole cell or a section of the cell can be studied in its 3-D state.
Investigation of whole cells under these powerful microscopes reveals that the cytoplasm, apart from its various cell structures, is made up of a network of fine protein filaments of 10-20 nm thickness. These filaments are associated with microtubules and microfilaments. Such a net work which pervades the whole cell is called microtrabacular lattice.
This structure is believed to be made up of tubulin like proteins. It is also associated with actin, microtubular associated proteins (MAPS) and other hundreds of unknown proteins. Their origin from microtubule organizing centers (MTOC) strongly indicates that it is made up some microtubular proteins. This structure undergoes depolymerization at 0 C0 and it requires ATP for its organization; furthermore Ca2* ions play an important regulatory role in the assembly and disassembly of these proteins. All these strongly favor the view that the microtrabacular network is made up of tubulins and its associated proteins. These filaments are also in contact with microtubular cytoskeleton as well as microfilaments. Association of polyribosomes at the intersection of this network is another important feature of this structure.
As the microtrabacular network exhibits dis-assembly and assembly, as it also undergoes contraction and expansion, as well as gliding movement all among the microtubular filaments, it is believed that they have an important role in the transportation of materials from one region to another region. This network of microtrabacular skeleton along with microtubules provides the frame work for the maintenance of the 3-dimensional shape of the cell. It is too early to speculate anything more than what has been observed.
But centrioles and its associated structures found in animal cells may help in protoplasmic streaming movements and they are also responsible for sol-gel transformation. Added to this they also help in determining the shape of the cell, movement of the cell, and transportation of the cytoskeleton structures play a very important role in the direction of cell division and differentiation. Many of the components also help in chromosomal movement and cytokinesis. However, the existence such structure is still enigmatic and some people don’t believe such structures exist. https://www.ncbi.nlm.nih.gov/.
NUCLEUS: Nucleus is the most important organelle of the cell and it plays vital roles in inheritance and gene expression, can be called as the Heart of the Cell. With the exception of bacteria and blue-green algae which contains all other organisms well organized nucleus. Such organisms are called Eukaryotes. On the other hand, bacteria and blue-green algae are referred to as prokaryotes for they lack well organized nucleus and their genetic material is simply a nacked DNA associated with some regulatory proteins.
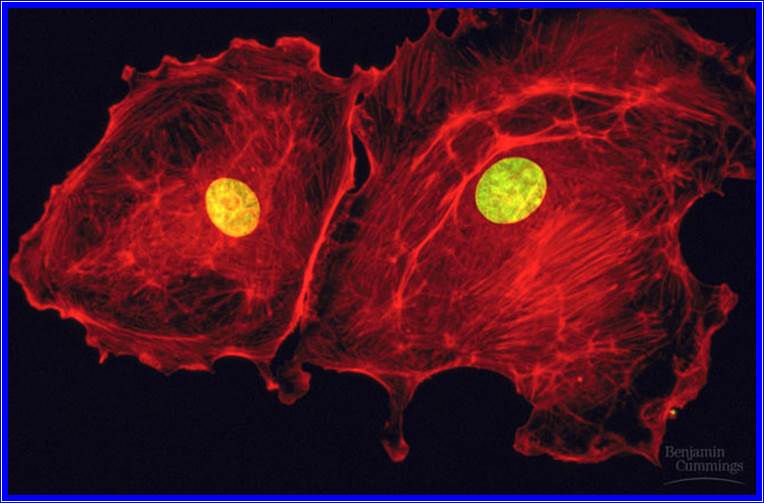
Shape, Size and Number: Most of the nuclei are spherical in shape, but variation in shapes like oval, bean shape, string shape, lobed and others is restricted to specific cell types. The size of the nucleus varies from 0.5 u to 2 u. Generally, the cell that is about to undergo cell division, particularly meiosis, contains a large nucleus. The number of nuclei per cell also varies. Cells with single nucleus are called monokaryotic and cells with two or more nuclei are referred to as dikaryotic and with many as coenocytic cells respectively. In some cases, like sieve tubes, in plants and red blood cells in animals, the nucleus disappears at the later stages of development. The position of the nucleus in the cell is not constant for it is displaced every moment by the sweeping movement of cytoplasm.
Structure-
Nuclear Membrane: Nucleus is bounded by membranes, within which chromatin threads and nucleoli are suspended in a fluid called nuclear sap or karyolymph. The nuclear envelope consists of outer and inner membranes each show unit membrane characteristics. The space found between these membranes is called perinuclear space and it is filled with fluid containing some granular and filamentous structures. The outer membrane shows continuity with endoplasmic reticulum. In fact, the outer surface of the outer membrane is studded with large number of ribosomes.
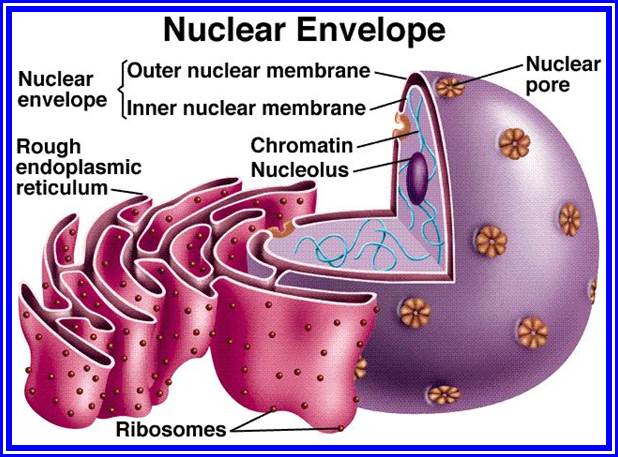
The nuclear membrane is not continuous but consists of number of pores, with a diameter of 50-150nm. The number of pores per unit area is not constant, and changes with the metabolic state of the cell. When cells are engaged in the synthesis of ribosomes in large numbers, the number of pores per unit area increases considerably and when it comes to a resting state, the number decreases. On the inner surface of the nuclear membrane, it is reinforced by intermediary skeletal proteins called lamins, which are associated with certain segments of chromatin.
Pore complex: The pores when observed under electron microscope show octagonal shape. At the periphery of the pore, both inner and outer nuclear membranes are in continuity. Furthermore, the pore consists of an octagonal shaped tubular structure called annulus. Each annulus is made up of eight granular protein subunits and they are arranged on either side of the pore (i.e. inside and outside of the pore) in octagonal fashion. Within the hallow of the annuli some amorphous material is found and it is in contact with the peripheral annular protein subunits by thin filaments. This entire structure is called pore complex. In its functions, it is not just a passive opening. It controls the movement of materials of various kinds and dimensions from nucleus to cytoplasm and vice-versa. As some of the proteins found in the pore complex exhibit ATPase activity, it presumed that the transport of materials is an active process. Inorganic ions, organic substances like amino acids, proteins (both small and large) are selectively transported into the Karyolymph; similarly, many nuclear products like, tRNA, mRNA, snRNA, and many noncoding (NC) RNAs and ribosomes are transported out through pore complex with ease. There are specific proteins act as importers (importins) and exporters (exportins).
Nucleolus: The nuclear sap consists of one or two electron dense regions, with granular and filamentous structures called Nucleolie. These may be spherical or ovoid in shape. The number is generally one, but two or more is not uncommon to find. Nucleolus is very important structure for; it acts as the site of synthesis and assembly of cytoplasmic ribosomes. Nucleolus is always found associated with secondary constriction region of one or more specific chromosomes. This region of the chromosome is also referred to as nucleolar organizer, because it is at this region the chromatin DNA, which consists of 150-500 tandem repeats, that code for r.RNA. This part of the nucleolus is often distinguished pars fibrosa, which consists of DNA and r.RNA strands. The clear liquid region found within and at peripheral region is called pars amorpha. The granular structures of various sizes, of which some are in the process of ribosomal assembly and some are fully formed, forms the region called pars granulose.
It is now known that the ribosomes are assembled with in the nucleolar region. The required r.RNA is synthesized in this region on ribosomal DNA segments and the ribosomal proteins are synthesized on cytoplasmic ribosomes, then they are transported into nucleolar region through pore complexes. These proteins then get assembled on various types of r.RNA and mature into functional ribosomal units. Such ribosomes are then transported out of nucleolus through pore complexes into cytoplasm.
Nuclear Sap or Karyolymph: The nuclear sap is relatively a dense liquid containing all the required components for DNA synthesis, RNA synthesis and other factors essential for their assembly into chromosomes. Many of the regulatory proteins are also found in this sap. The maintenance of various nucleotides pools and protein pools is important in the functioning of chromosomes.
Karyolymph contains all the required components for the DNA replication and repair, transcription, assembly of ribosomes, all the said components are imported from the cytoplasm. The nuclear sap also contains few unstructured regions, which can be stained, called cajal bodies, which contain coiled proteins called Coilins. These structures are involved in processing small nuclear RNAs called snRNAs. In recent years many more spec like structure is found within the nuclear sap and structure and functions are more revealing.
CHROMOSMES
Chromatin: Suspended within the nuclear sap are the net works of threads, when stained they take color; hence they are called chromatin threads (chroma-colour, tene-thread). Some of the threads, particularly their ends are associated with either pore complexes or inner nuclear membranes. Chromosomal threads are attached to a proteinaceous matrix at the inner surface of the nuclear membrane; thus, the positions of chromosomal threads are fixed. This interphase chromatin network is not a constant feature, but changes as and when the cell passes through various stages of cell division. Nevertheless, the chromatin at the G1 stage, appears to be diffused, thin, single stranded and coiled, but this single stranded chromatin threads undergo duplication to form double stranded chromatin threads at S-stage. During duplication chromosomal DNA replicates, and necessary histones and nonhistones are drawn from the nuclear sap to form sister chromatin threads. With the progress of interphase into prophase, the long, thin threads undergo a continuous process of condensation resulting in shorter and thicker chromosomes. At the same time, the chromatin distangles from the network and chromosomes slowly get resolved into individual threads.

E. micrographic Picture of Chromosomes
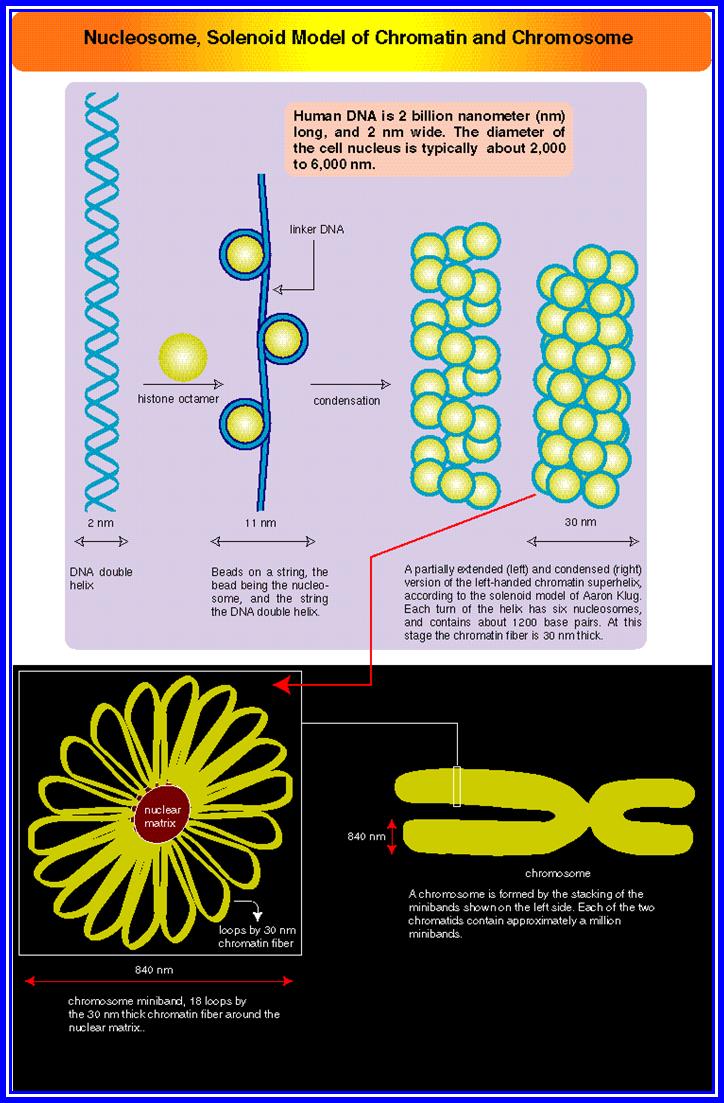
Nucleosomes- solenoid model of organization of chromatin Threads.
By the time, cells reach metaphase stage, chromosomes undergo maximum condensation, and individual chromosomes can be made out under normal microscope. At this stage the number and the detailed structure of them can be studied with light microscope or electron microscope.
Number of chromosomes: The number of chromosomes varies from organism to organism (2-1600) and this number is constant and characteristic for a given species. (Table below).
|
Common name |
Specific name |
Chromosomal number(2n) |
Fruity fly |
Drosophila |
8 |
|
Frog |
Rana pipiens |
20 |
|
Gorilla |
Gorilla gorilla |
48 |
|
Monkey |
Macaca mulatta |
42 |
|
Man |
Homo sapiens |
46 |
|
Garden pea |
Pisum sativum |
14 |
|
French bean |
Phaseolus vulgaris |
14 |
|
Onion |
Allium cepa |
16 |
|
Cabbage |
Brassica oleracea |
18 |
|
Coffee |
Coffea arabica |
44 |
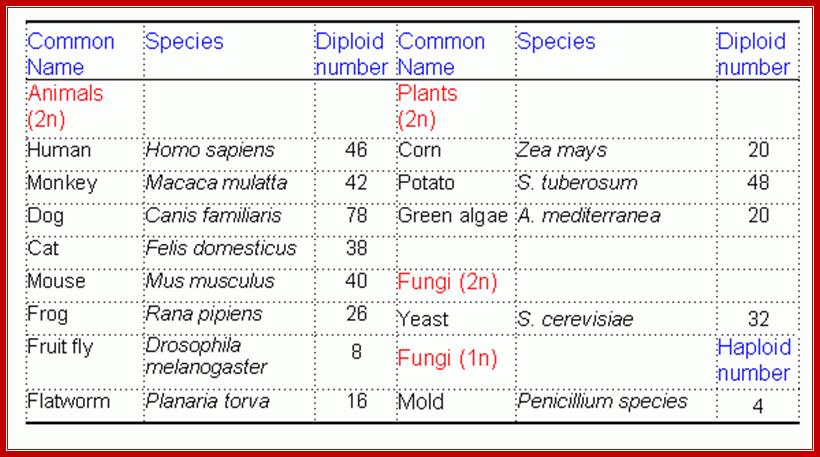
The number of chromosomes is denoted by the terms Karyotype which may be either haploid or polyploid. The haploid karyotype consists of one set of chromosomes, where every individual chromosome is structurally and genomically different from the others and exhibit unique characteristics. For example: in the case of onion, the haploid (n) chromosome number is 8 and let us call them as A, B, C, D, E, F, G, H. Here, each chromosome is different and unique in its genomic content. And such a set of chromosomes is called haploid set. If two such sets of chromosomes are put together in the same nucleus then that nucleus or the organism that posses it, is called diploid i.e. here two haploid sets are present, they are called homologous chromosomes or homologous pairs. On the other hand, as A and B chromosomes are being different, they are called non-homologous chromosomes. The terms triploid (3 n), tetraploid (4 n) and polyploid just indicates the number of sets present in the nucleus.

A group of organisms belonging to a particular species, though show a constant chromosomal number, say diploid, they often exhibit variation in chromosome numbers, either by loss or gain of one or more chromosomes. In some cases, the entire set of chromosomes may be involved. This variation in chromosomal number leads to variation in the morphology and functional behavior of organisms. Such changes may ultimately lead to variation and origin of species, provided they survive. This is one of the fundamental steps in organic evolution.

Size of chromosomes: Chromosomal size varies from organism to organism; however, a particular size of chromosomes is constant for a given species. Some of the plants of Cyperaceae and Luzula have very small chromosomes, but plants like Trillium have quite large chromosomes of the size 30µ in length. However, in a given karyotype, all the chromosomes are not of the same size (asymmetrical karyotype) and rarely do we find organisms with chromosomes of the same size (symmetrical karyotype). Some chromosomes like salivary gland chromosomes (Drosophila), Lampbrush chromosomes, (Xenopus laevis) and chromosomes in the endosperm haustoria of Phaseolus are 100-1000 times larger than their somatic chromosomes. These are called special type of chromosomes or call them as ‘Giant Chromosomes’. They become so because of the necessity. Here each of the genes is duplicated to thousand times and very helpful in producing transcripts in large numbers for developmental purpose.
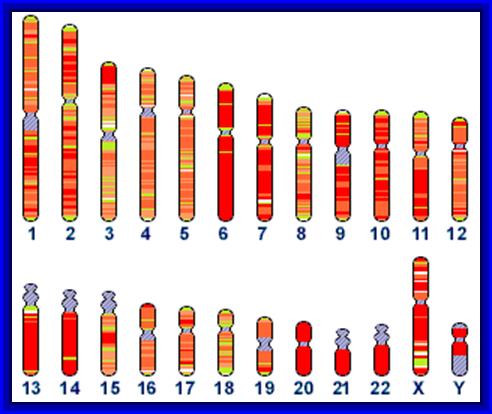
Human chromosomes; 1to 22+X/Y


Chromosomal bands-heterochromatin and euchromatins and size of chromosomes-in Mbps
Shape of the Chromosomes: Almost all chromosomes look like spirally coiled thread, but during cell division particularly at anaphase stage, chromosomes show a specific bent shape. This is due to the position of primary constriction or centromere. Accordingly, the chromosomes are called Metacentric (V-shape), Sub-metacentric (J-shape), Acrocentric (rod shape),
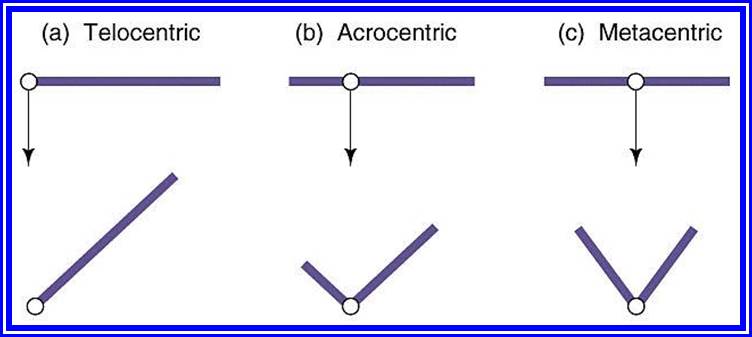
Telocentric (rod shape), Acrocentric, Metacentric -not shown; (Polycentric (wave shape) and Diffused (rod shaped but move horizontally).
Sex chromosomes and Autosomes: Higher organisms like man, monkeys and some plants where male and female sexes are morphologically differentiated, the cells in them contain two types of chromosomes, called Autosomes and sex chromosomes. The latter classification is based on the X and Y chromosomes. This classification is based on the chromatin nature and function. For example: human beings (Homo sapiens) have 46 chromosomes of which 44 are autosomes and 2 are sex chromosomes. Autosomes are believed to control the development of somatic body and sex chromosomes are responsible for the expression of sexual organs and characters. If two XX chromosomes are present, it determines the female sex of the organism and its related characters are expressed; if one X and one Y chromosomes are present, male character is expressed. The X chromosome is considered to express female character and Y and the male character. The X chromosomes are more or less euchromatic and Y are heterochromatic. Furthermore, the sex expression varies in different organisms. All in all, it is the interaction between autosomes and sex chromosomes that ultimately determines the expression of sexes through the mediation of specific hormones like estrogens, female hormones and Androgens male hormones.
Chromosomal structure;
Using optical microscopes, if chromosomes of metaphase (at which chromosomes are at maximum condensation) are observed, chromosomes appear to be double stranded threads, which are relatively coiled to each other. These threads are referred to as chromatids or chromonemata. If such chromatids are carefully observed under high resolution light microscope (2000 times enlarged), each of them appears to be spirally coiled with apparent gyrations.

Pattern of chromosome condensed or diffused chromatins (above diagram).
Many chromosomes show differential condensation, because of which some parts take more stains and other less stain. The former is called heterochromatic segments and the later are euchromatic. This differential staining behavior is called heteropycnosis. The heterochromatic segments may be either tightly condensed (take more strain).
Less condensed (takes less stain), this feature is called as positive heteropycnosis and negative heteropycnosis respectively. In some cases, the entire chromosome appears to be heterochromatic because of greater condensation. The heterochromatin can be constitutive as in CEN and telomeric regions and it can be facultative also in other regions; the position of such constitutive regions does not change but facultative heterochromatin may change during development and from one tissue to the other. Formerly, these heterochromatic regions were believed to be genetically inert, now they are known to contain genes and they do express. However, now, constitutive heterochromatin regions like CEN and telomeres are know to contain highly repetitive DNA segments.
The inheritance of Mendelian factors or characters through chromosomes was not substantiated till the discovery of chromosomes. This has, however, led to unit character inheritance, specially located within chromosomes. These unit characters are now called genes. Nevertheless, the problem of genes which are arranged in chromosomes was an enigma, but the discovery of bead like structures in meiotic chromosomes, called chromomeres, has given an impetus in unraveling this problem. Meiotic chromosomes particularly at leptotene stage appear as fine strings of beads.
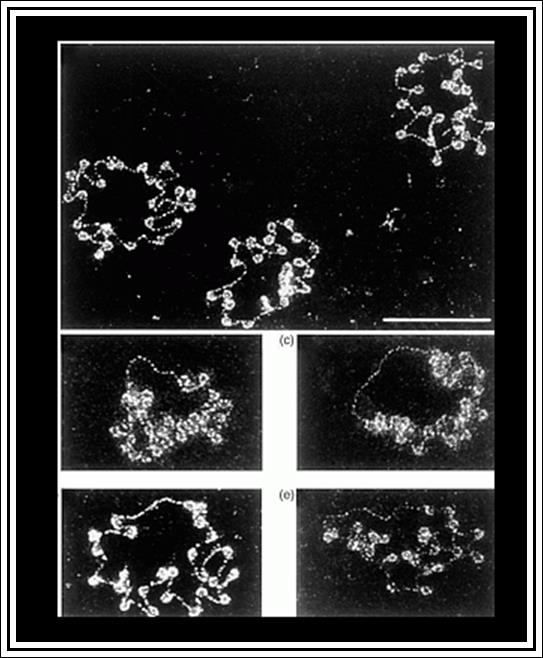
Chromomeres- Pattern
These beads like structures were called as chromomeres and were equated to individual genes, organized into bead like structures. Such chromomeres were assumed to be held by nongenetic threads. But later, chromomeres were found be none other than the coiled expressions of chromonemata; when two ends of such chromatids are stretched apart, the chromomeres disappear. Nevertheless, the concept of linear arrangement of genes in chromosomes has been accepted. Morgan and later Hunt’s cytogenetic experiments have furthered the concept of gene as a unit of heredity, a unit of recombination, a unit of mutation and a unit of function and chromosomes form unit of linkage.

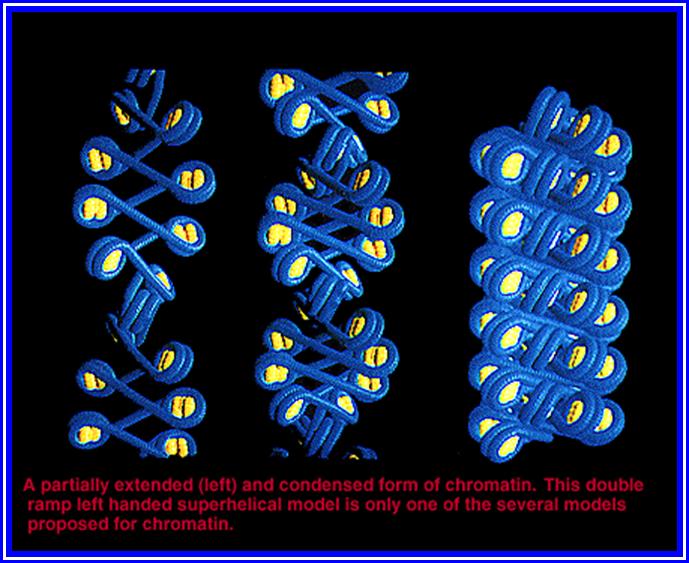
Morphologically the metaphasic chromosomes appear to be simple, coiled threads of uniform thickness; here and there the chromosomes contain constricted or narrow regions. These are called primary constrictions and secondary constrictions respectively. The primary constrictions and secondary constrictions are further differentiated and characterized by their behavior and functions.
Centromere: The primary constriction is also called centromere for it is the region at which chromosomes get attached to tractile fibres and they lead the anaphasic chromosomal movements. If the centromere is destroyed by direct X-radiation hits, chromosomes behave abnormally and lose their directional movements. This results in loss or gain chromosomes at the end of cell divisions. Thus, centromere appears to be a non stainable gap in the chromosomal thread. However, in metaphasic chromosomes, though the arms are double stranded, the centromere still appears to be single stranded, but in actuality it is double stranded but held tightly by proteins called adhesins.
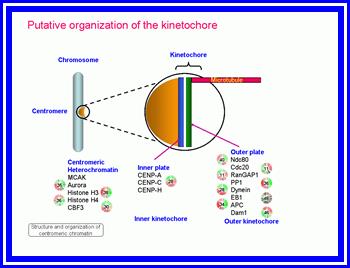
C-shaped structure on either side of the centromere.

Nature-Reviews-Genetics.
In a holocentric chromosome, the entire chromosome acts as the centromere. Although this sort of chromosome is not typical, it does occur in certain types of organisms, that a biologist is likely to meet. For example, in nematodes (the nematode Caenorhabditis elegans is widely used in research).

The centromere kinetochore region:
At the heart of the kinetochore is a specialized nucleosome that contains centromere protein (CENP)-A, a histone H3 homologue. Several inner kinetochore components (cyan and purple ovals) associate with kinetochores throughout the cell cycle. Many other proteins are recruited to the outer kinetochore specifically in mitosis. They provide a landing platform for the spindle-assembly checkpoint (SAC) proteins. The Ndc80/HEC1 complex seems to be directly involved in microtubule binding. Several microtubule-plus-end-binding proteins (+TIPs) are important for microtubule to kinetochore attachment. Most proteins indicated in this drawing are present at kinetochores in all metazoans. This image is linked to the following Scitable pages:
https://www.nature.com/
Recent electron microscopic studies clearly show that on either side of the centromeric region, in line with the chromosomal arms, it is covered by ‘C’ shaped structure called Kinetochore. Thus, mitotic chromosomes contain two kinetochore structures are present at each centromere. Each kinetochore is made up of three regions. The outer most region is cup shaped structure called commissural cup and it is relatively thick, its outer surface large number of processes called corona are present. Amidst the corona a number of (3-10) microtubules are found to be penetrated as deep as to reach chromonema. The middle region is less dense, but the inner region is dense and it is in contact with centromeric chromonema. However, the entire kinetochore structure appears to be a highly specialized region and it is likely to participate as the nucleating centre in the polymerization of tubulin into tractile fibers during the formation of mitotic apparatus.
Another interesting feature of the centromere is that, on both flanking regions of the centromere the chromatin material is heterochromatic. And it is now known that the DNA in this region is highly repeated (highly repetitive class of DNA; however, their function is not known.
Secondary constriction or Nucleolar organizer: Similar to that of primary constriction another constriction is present only on specific chromosomes. Such constriction is called secondary constriction and it is characterized by its nucleolar formation; hence it is also referred to as Nucleolar organizer. This region contains DNA segments for ribosomal RNA. The genes present, here, are redundant or tandemly repeated. In some cases, like frog oocytes these genes get amplified at the end of the telophase. The DNA in the region opens out, and starts transcribing ribosomal RNA as large precursors. Later this RNA is sliced and processed into 18s 28s and 5.8s RNAs. These RNAs in turn get associated with different ribo-proteins sequentially and functional ribosomes are produced. If the secondary constriction segment of the chromosome is knocked off, the nucleolus fails to appear and the cells die prematurely. In human chromosomes the nucleolar organizers are found on chromosomes 13, 14, 15, 21 and 22.
SAT Chromosomes: Short chromosomal segments, at the terminal region contain blob like structure, beyond secondary constriction; they are called Satellites. Such chromosomes are called SAT chromosomes. Heitz, who coined this term, is in the opinion that these segments are lacking in thymidilic acid, i.e., Sine Acido Thymidine (SAT). Otherwise, this region, as it is lacking Thymidilic acid, it is rich in Guanidilic and Cytidilic acids. Whether such GC rich heterochromatic chromosomal segments contain some redundant genes or not, is not known. They are rich in short tandem repeat DNA segments. Their function is also a mystery.
Telomeres: In all most all organisms, with out any exceptions, chromosomal ends have heterochromatin materials. Such structures are referred to as telomeres. The presence of such structures is found to be non-sticky and prevent the attachment of broken chromosomal ends in wrong way. If telomeres are cut off, the ends become sticky. However, the recent in situ hybridization techniques have demonstrated that heterochromatic telomeres contain repetitive DNA of particular sequence such as GGGATT. The telomeric DNA runs many thousands of base pairs of such repeats
Ends of chromosomes-called Telomeres
The DNA analysis of such ends show, the 3’end of the DNA; these regions are free and can be subjected to exonuclease action. So, in order to protect such ends, in this region the DNA is organized into loop like structure associated with a variety of proteins to protect from exonucleases. Even the replication of this telomeric DNA uses a short RNAs as the primers and reverse transcriptase for generating telomeric DNA. However, the size of telomeres changes; they may expand or contract. Damaged telomeric ends can cause or induce cancer.
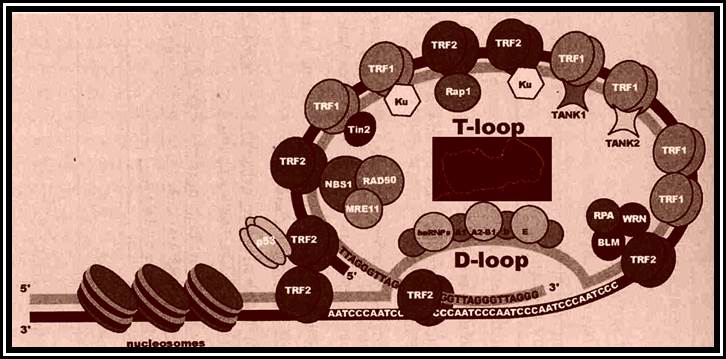
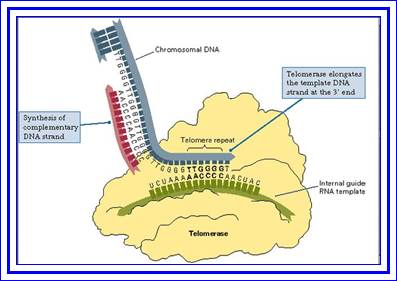
Showing Telomeric ends at molecular level.
Ultra structure of Chromosomes
Observations of metaphasic chromosomes under optical microscopes of reasonable resolution, shows them as fine, plectonemally coiled threads. Beyond this, it is difficult to understand the internal organization of chromosomes. Nevertheless, the techniques, like whole mount chromosomes combined with biochemistry and electron microscopy, have revealed that chromosomes are made up of highly folded but coiled chromonemal threads of 250-300 A0 thickness, with ends visible nowhere.
Biochemical combined with molecular studies studies has revealed that the chromosomes contain DNA, histone and non-histone proteins. The complexity of the organization of the above said components has further compounded with the discovery of various components of histones and nonhistones. Histones are found to be basic proteins and different kinds such as H1, H2A, H2B, H3, and H4. These proteins are rich in lysine and Arginine residues that are the reason they show basic properties.
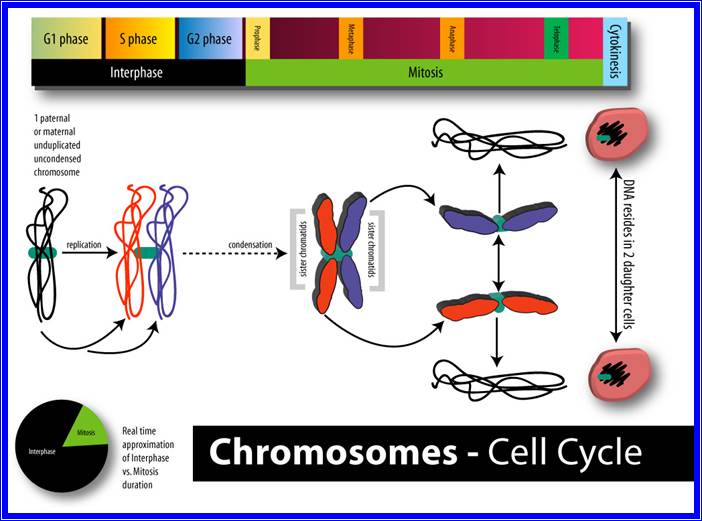
Chromosomal structural pattern at different stages of cell division;
The nonhistone proteins are found to be acidic in nature and 100-120 or more different kinds have been identified by 2-D gels.
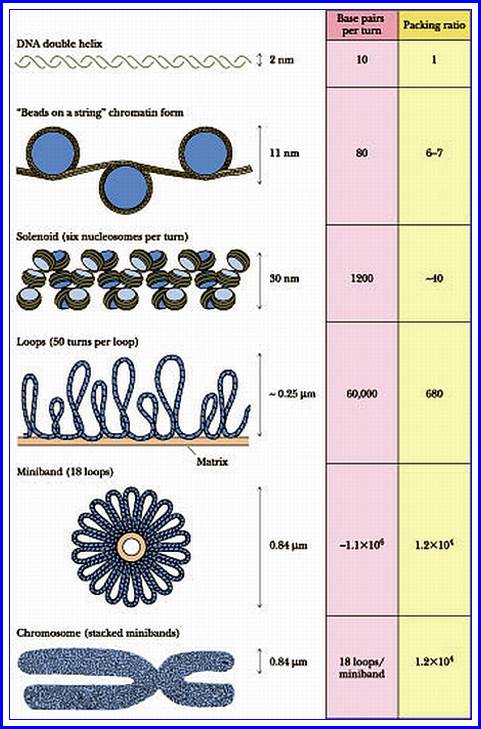
When chromatin is added to a salt medium, the chromatin threads loosen and spread out into fine bead like structures called Nu-bodies or Nucleosomes. The Nu-bodies were further identified as to contain histones as octamer (2H2A 2H2B, 2H3 and 2H4) and DNA double helix of about 140 base pairs length is coiled around it. The nucleosomes are the fundament chromonemal units, where each of the histone as tetramers are superposed one on each. Each of the histones contain a folded structure with free N-terminal tails and H2A contains a C-terminal tail too. They play very important role in condensation of the chromonema and decondensation and differential gene expression. The amino acid sequence in the tail provides a kind of signatures such can utilized as Chromodomain and Bromodomain, very very essential gene expression or repression. The DNA thread found in between two such Nu-bodies is called linker DNA which consists of 50 to 100 base pairs.
Furthermore, the electron microscopic studies of polytene chromosomes reveal that the Nu-bodies or nucleosomes are biconvex disc shaped structures with histone octamer as the core, around which DNA coiled 1 ľ times. These structures have been recognized as the fundamental units of chromosomal fibres.
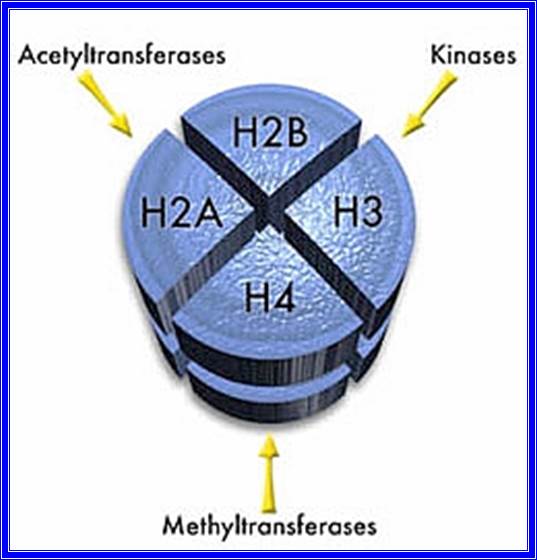
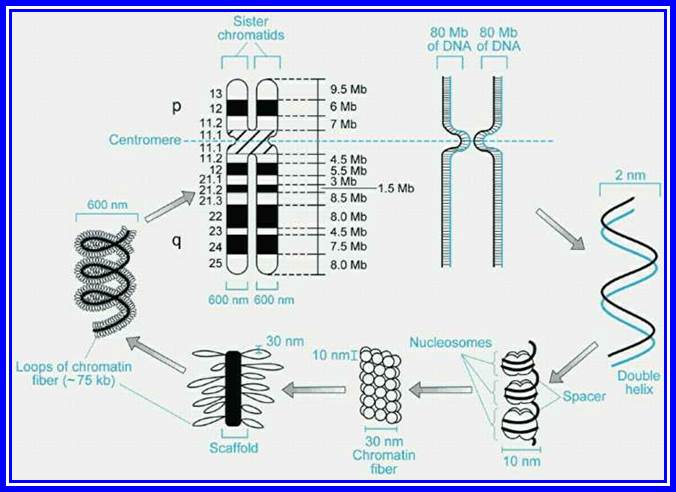
Such fundamental chromosomal fibres with Nu-bodies as units, undergo solenoid coiling or produce compact zigzag conformation in the presence of H1 proteins. This forms the basic thread of chromosomes called chromonema of 30nm size. Each of the solenoid or zigzag structures may contain about 6-7 nucleosomes. Then this solenoid thread, in association or in combination with specific proteins, undergoes further compaction with emanating loops from the scaffold. The DNA in the loops of different sizes are attached to scaffold proteins by a specific DNA sequences called MARs, called so they are matrix attachments regions. The size of the loops varies in their structure and dimensions; the dimensions can be of 300A size. These chromonemal compact structures are visible under light microscope. These threads show a contraction of 1300 to 1500-fold to that of string of beads.
Artificial, these are man made synthetic chromosomes, look at the arrow pointing to an artificial-synthetic chromosome.
Such chromonemata are further coiled in metaphasic chromosome which exhibits a 6 x 103 – 7 x 103-fold condensation. However, the coiling and condensation of Nu-bead fibres into microscopically visible chromosome, is aided and augmented by a number of non-histone proteins as binding factors to form that can be observed under optical microscopes. If such chromosomes are subjected to histone digestion and dispersed under certain detergent cum salt solution, the histones are selectively removed, but retain the non-histone proteins intact. All the histone free DNA molecules spill out in the form of loops of fibers of various sizes.
These and other studies, notwithstanding the complexity of the association of DNA with proteins, have shown that each chromonemata is made up of single, but a long DNA molecule, which supports the concept that the genes are arranged in linear order, where a segment of DNA acts as the unit of heredity. Such chromosomes go through condensation and relaxation during cell division in stage specific manner. In interphase most of the chromosomes are relaxed and bound to inner lamina proteins and found in fixed positions.
In recent years, with greater understanding of the structure and functions of chromosomes, molecular biologists have created chromosomes in the lab, they can be stained and observed along with others normal ones. This will provide great benefits to cure certain hereditary diseases, I am sure not immediately, it may take few more years.
Giant chromosomes:
Chromosomes, in most of the organisms show a cycle of condensation and decondensation during various stages of cell divisions. But in some organisms, chromosomes enlarge considerably to the tune of 250-500 times the normal somatic chromosomes. The difference in their sizes can be observed under optical microscopes with lower resolution. These are called giant chromosomes and they are not just restricted to one species but found to occur in various organisms like insects, frogs, salamanders and even plants.
However, their occurrence is restricted to certain stages in the life cycle. Two such giant chromosomal types’ i.e. salivary gland chromosome and Lampbrush chromosome have been extensively studied.
Salivary gland chromosomes or polytene chromosomes: Salivary gland chromosomes are found in salivary gland cells of 11th day larvae of dipteron class of insects. They are present in the larvae of Chironema, Drosophila melanogaster, mosquitoes and other insects of the same class. Such giant chromosomes are also found in the endospermal haustoria of Phaseolus vulgaris.
In the somatic cells of the insect, these chromosomes remain normal in their size, but on hatching of eggs and when the larva reaches the 11th day and is about to undergo metamorphosis into pupa, the somatic chromosomes found in the salivary glands undergo a dramatic change in the size and activity. At this stage of development, chromosomes undergo repeated chromosomal DNA replication without separation, which leads to the multiplication of two chromonemal threads into 1000-1084 strands.
Show chromatin condensed and diffused regions.
Furthermore, these somatic homologous pairs of chromosomes are found to be in pairs or synaptic state.

Structurally these chromosomes contain thousands of genes longitudinally oriented and the chromonemal threads arranged parallel to each other. These threads also show cross banding of various sizes. Some bands take greater stain and others stain less. The Darkly stainable bands are heterochromatin segments where the chromosome is densely packed with chromatin material.
Occasionally some of these darker bands containing highly condensed chromatin or chromatin in the inter bands open out into nacked DNA loops and same are transcribed, and one can find RNAPs associated with transcribed m. RNAs. Even some inter-band regions also open out. The transcriptional activity has been identified by the use of radioactive precursors. This region represents intense gene expression activity. Because of this reason, this region appears be puff like structure which consists of opened out DNA strands in the form of a ring like structures called Balbiani rings. In each loci thousands of loops of various sizes can be seen, so appear as puff like structures. Such puffs can be induced in the chromosomes by applying an insect hormone called ecdysone. This hormone induces the transformation of larva into pupa, during which many morphological structures and functional activities of the larva undergo dramatic changes. Probably, because of these reasons, the 11th day larval chromosome undergoes differential gene activation to synthesize required protein products for the metamorphosis in stepwise manner. During the course development many of the genes expressed earlier, get silenced and new set of genes get expressed in a form called cascading, that is one leads to the other, a differential expression required for the metamorphosis. Many of the genes expressed during these stages have been identified.
Ultra-structural features of Lamp brush chromatin.
https://www.nature.com/scitable/
Lampbrush chromosomes: Chromosomes which appear as bottle brush or the brush that is used to clean the lamp glasses are often called Lampbrush chromosomes. These chromosomes are highly elongated (5900 um) and they are found in the oocyte’s frogs. Here during the development of oocytes, the homologous pairs of chromosomes undergo into extended threads by the uncoiling of specific chromomere and differentiation. To begin with, the homologous pairs undergo synapsis, and later they are held to each other at chiasmatic regions. Each homologous chromosome consists of two chromonemates containing a large number of granular structures called chromomeres. Many of these chromomeres, all along the length of chromatin threads open out in the form of large lateral loops of nacked DNA. Soon, these loops will be covered by a matrix, which is made up of a pool of RNAs and RNA polymerases.
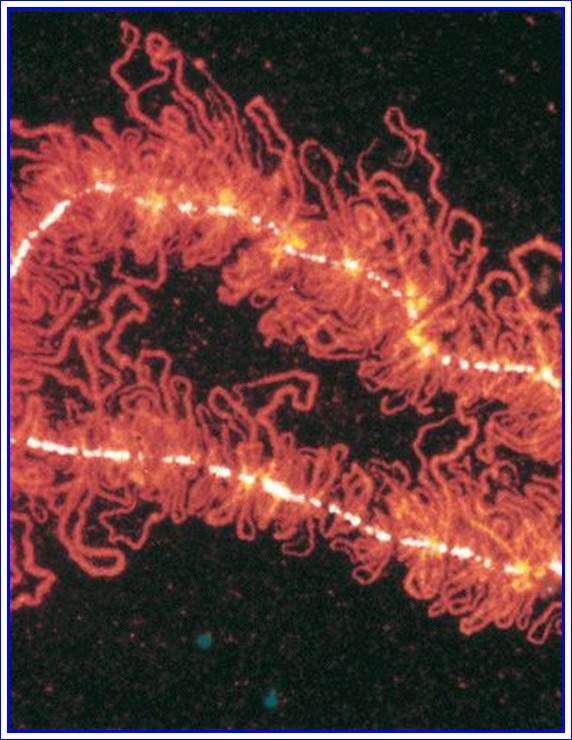
Lamp brush chromosomes.
These lateral loops now start intense transcriptional activity, with the result, innumerable RNAs are synthesized. Each loop consists of many genes of the same kind or different separated by noncoding regions called spacer DNA.
Transcriptional activity is initiated at several sites simultaneously by the binding of RNA polymerases and soon RNA strands are formed with the progression of RNA polymerase. As soon as the initiating site is free, another RNA polymerase settles and starts synthesis at various stages and each of these segments appears as the branch of a christmas tree. RNA synthesis is required for the developing egg and many of the poly A-RNAs (mRNAs) are stored to be translated only when the egg is fertilized. This is another instance of differential gene activity, required for certain development process.
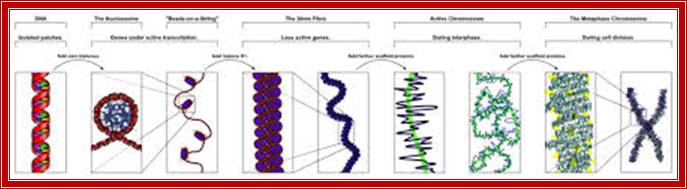
Chromosomes- WIKIPEDIA.



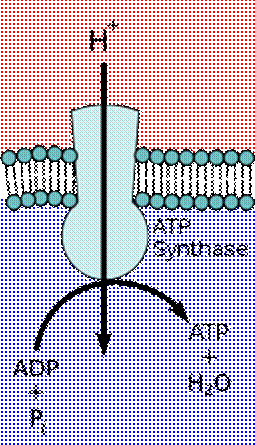



![LUCA systems and environment[10]](1_CELL_STRUCTURE_files/image009.gif)

